Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
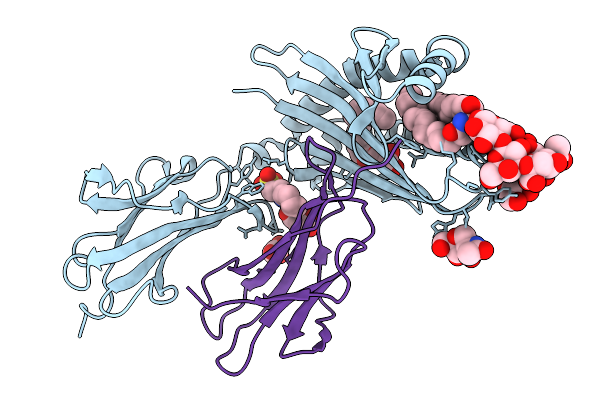 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LIPID BINDING PROTEIN Ligands: NAG, A1CB5, KZF, MLA, A1CB4 |
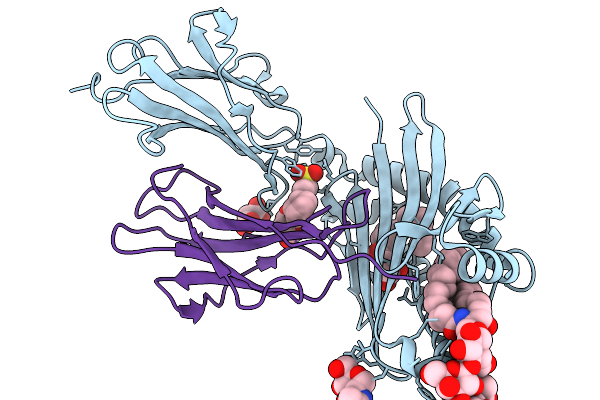 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LIPID BINDING PROTEIN Ligands: NAG, A1CB5, KZF, MLA, CL, A1CB6 |
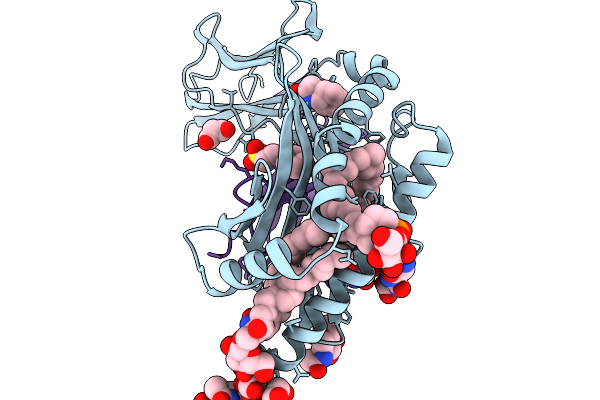 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LIPID BINDING PROTEIN Ligands: NAG, MK0, GGG, KZF, MLA, A1CB7 |
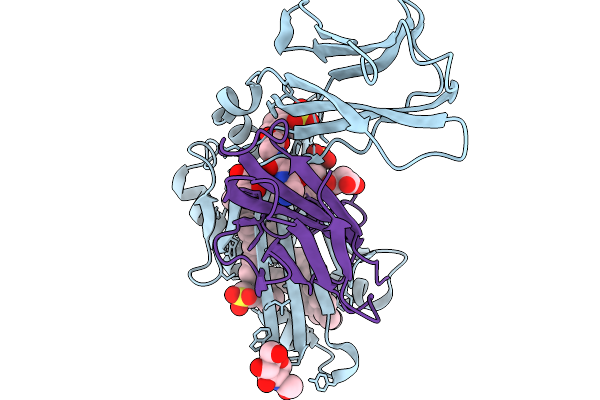 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LIPID BINDING PROTEIN Ligands: Z7B, NAG, D12, KZF, GGG, MLA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: DE NOVO PROTEIN Ligands: 2PE, PGE, MLA |
 |
Crystal Strcture Of Adenosylcobinamide Kinase/Adenosylcobinamide Phosphate Guanylyltransferase Cobu From Akkermansia Muciniphila, Crystal Form I
Organism: Akkermansia muciniphila atcc baa-835
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: MLA |
 |
Crystal Structure Of An Engineered Enzyme Containing A Genetically Encoded Thiourea, Ct1.5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: BIOSYNTHETIC PROTEIN Ligands: MLA, ACY, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ONCOPROTEIN Ligands: GDP, MG, FMT, MLA, ACT |
 |
Organism: Albifimbria verrucaria
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: CU, OXY, GOL, PGE, MLA, CL |
 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum, Wild Type Semet Derivative (Lagh157)
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: ACT, GOL, MLA |
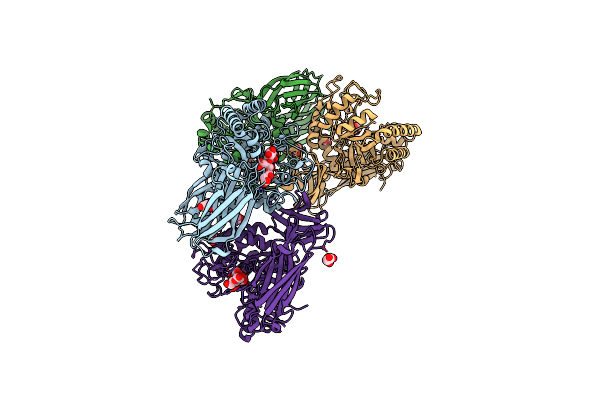 |
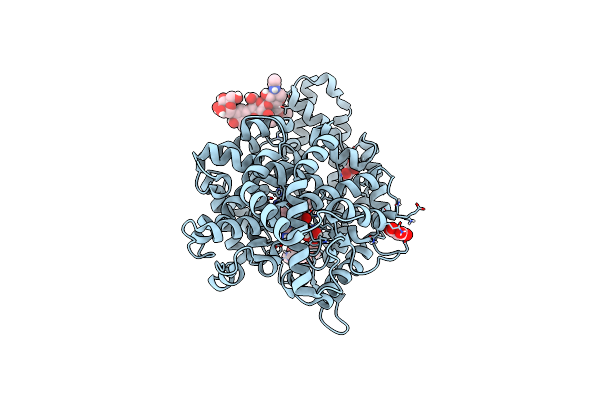
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) In Complex With Laminaribiose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: MLA, GOL, BGC, PG4 |
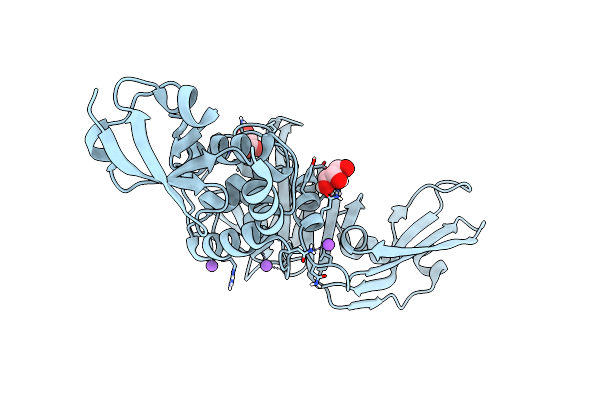 |
Sars-Cov-2 Papain-Like Protease (Plpro) C112S-K191D-Q222D-K229R-Q230R-C271S Mutant
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: MLA, GOL, NA, CL |
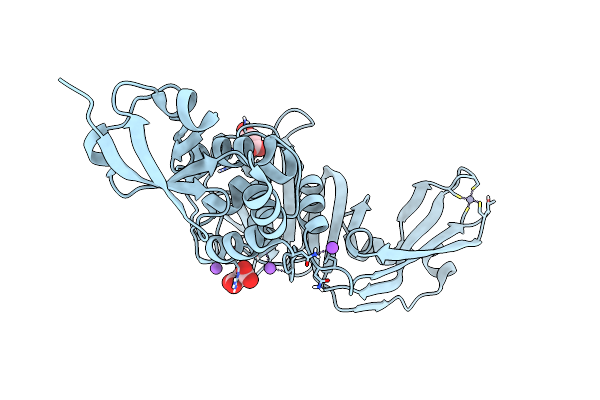 |
Sars-Cov-2 Papain-Like Protease (Plpro) C112S-K191D-Q222D-K229R-Q230R-C271S Mutant
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: ZN, MLA, NA, CL |
 |
Crystal Structure Of Angiotensin-1 Converting Enzyme C-Domain In Complex With Dual Ace/Nep Inhibitor Ad016
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-04-16 Classification: HYDROLASE Ligands: NAG, BO3, MLA, ZN, CL, A1IRS |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-03-05 Classification: TRANSFERASE Ligands: GTP, FMT, MLA, GOL, GDP, ACT |
 |
Q108K:K40L:T51V:T53C:R58Y:Y19W:Q38L:Q4A:T29L Mutant Of Hcrbpii Bound To Fr1V And Irradiated With Uv Light At Ph 7.6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-02-12 Classification: RETINOL BINDING PROTEIN Ligands: ZFJ, ACY, MLA, GOL |
 |
Organism: Metagenomes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: MLA, BGC, NHE |
 |
Organism: Metagenomes
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: NHE, NA, MLA |
 |
Crystal Structure Of Gh1 Beta-Glucosidase Td2F2 E352Q Laminaribiose Complex
Organism: Metagenomes
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: MLA, NHE, SO4 |
 |
Ests1 Phthalate Ester Degrading Esterase From Sulfobacillus Acidophilus In Complex With Jeffamine
Organism: Sulfobacillus acidophilus dsm 10332
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, JEF, MLA |