Search Count: 23
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-19 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, MHA, SO4 |
 |
Crystal Structure Of Leukotoxin Luke From Staphylococcus Aureus At 1.9 Angstrom Resolution
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-04-06 Classification: TOXIN Ligands: P6G, MHA, SO4 |
 |
Crystal Structure Of Human Complement C5 In Complex With The K8 Bovine Knob Domain Peptide.
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-03-10 Classification: IMMUNE SYSTEM Ligands: EOH, MHA, MPD, CYS, TAM |
 |
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Nitd-349
Organism: Mycolicibacterium smegmatis mc2 155, Enterobacteria phage rb59
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-12-02 Classification: MEMBRANE PROTEIN Ligands: FFU, L6T, MHA |
 |
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Spiro
Organism: Mycolicibacterium smegmatis mc2 155, Enterobacteria phage rb59
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2020-12-02 Classification: MEMBRANE PROTEIN Ligands: L6T, MHA, FG0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-02-19 Classification: HYDROLASE Ligands: HFH, GOL, NA, MHA |
 |
Cathepsin-K In Complex With Amino-Oxaazabicyclo[3.3.0]Octanyl Containing Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-02-19 Classification: HYDROLASE Ligands: MHA, J6B |
 |
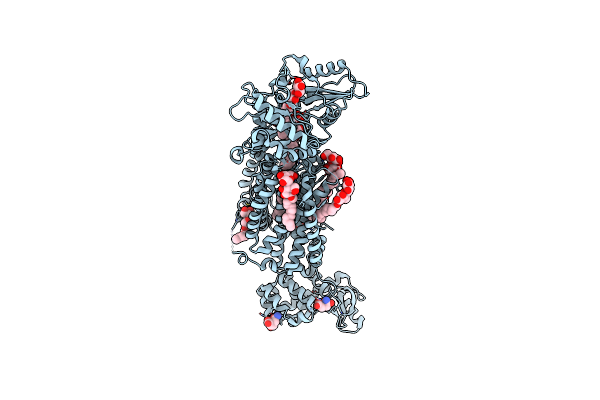
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-12-26 Classification: MEMBRANE PROTEIN, HYDROLASE Ligands: MHA, L6T |
 |
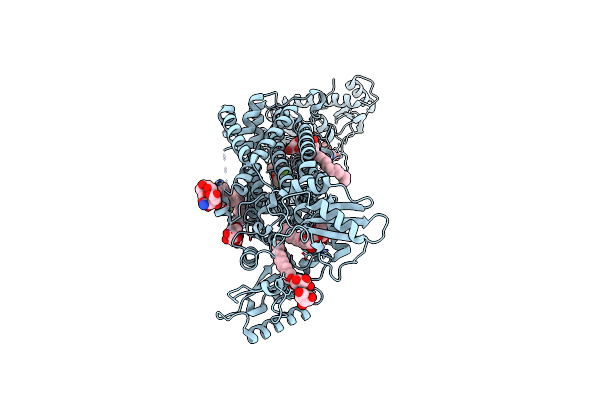
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Sq109
Organism: Mycobacterium smegmatis str. mc2 155, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-12-26 Classification: MEMBRANE PROTEIN, HYDROLASE Ligands: 3RX, LMT, MHA |
 |
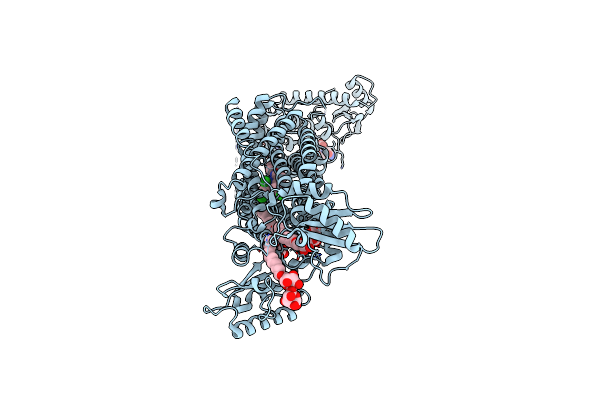
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Au1235
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2018-12-26 Classification: MEMBRANE PROTEIN, HYDROLASE Ligands: 9ZF, L6T, MHA |
 |
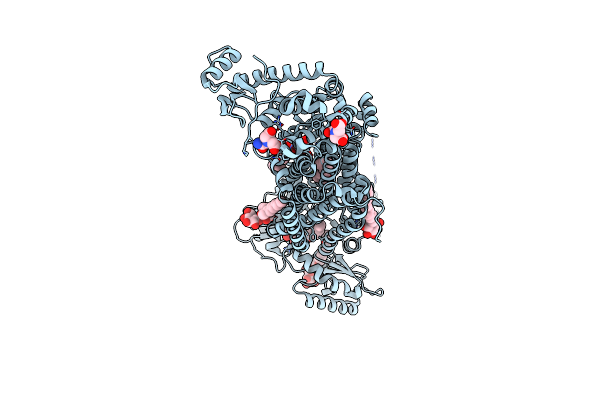
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Rimonabant
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-12-26 Classification: MEMBRANE PROTEIN, HYDROLASE Ligands: AY6, L6T, MHA |
 |
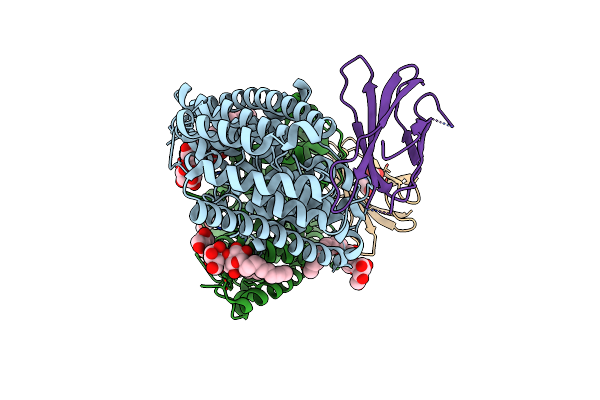
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Ica38
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2018-12-26 Classification: MEMBRANE PROTEIN, HYDROLASE Ligands: MHA, L6T, J9E |
 |
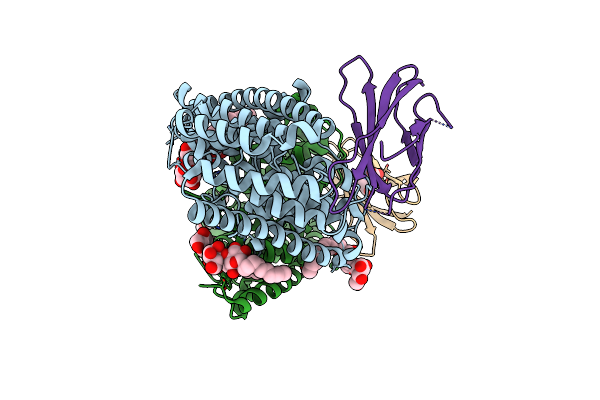
Organism: Enterococcus casseliflavus (strain ec10), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-07-04 Classification: TRANSPORT PROTEIN Ligands: DMU, F, MHA |
 |
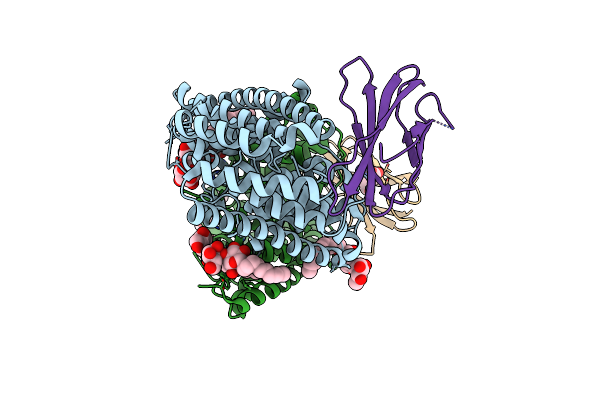
Organism: Enterococcus casseliflavus (strain ec10), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2018-07-04 Classification: TRANSPORT PROTEIN Ligands: DMU, F, MHA |
 |
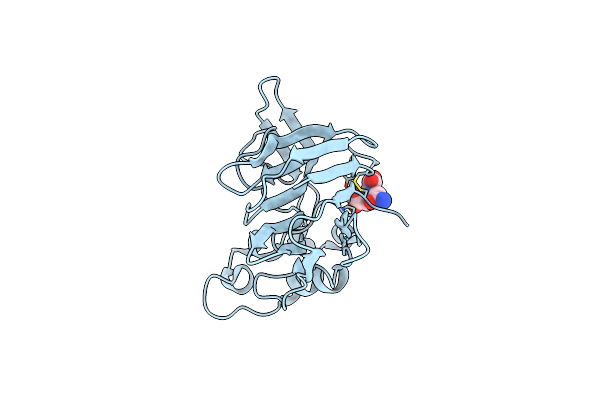
Organism: Enterococcus casseliflavus (strain ec10), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2018-07-04 Classification: TRANSPORT PROTEIN Ligands: DMU, F, MHA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-08-09 Classification: PLANT PROTEIN Ligands: MHA |
 |
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2017-08-02 Classification: HYDROLASE Ligands: CA, MHA |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: MG, SO4, MHA |
 |
Structure-Function Analysis Of Functionally Diverse Members Of The Cyclic Amide Hydrolase Family Of Toblerone Fold Enzymes
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: SO4, MHA, NA, CL |
 |
Structure-Function Analysis Of Functionally Diverse Members Of The Cyclic Amide Hydrolase Family Of Toblerone Fold Enzymes
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: CL, MHA, NA |

