Search Count: 29
 |
Organism: Moloney murine leukemia virus (isolate shinnick), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-02-27 Classification: HYDROLASE/DNA |
 |
Organism: Moloney murine leukemia virus (isolate shinnick), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-02-27 Classification: HYDROLASE/DNA |
 |
Organism: Moloney murine leukemia virus (isolate shinnick), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-02-27 Classification: HYDROLASE/DNA |
 |
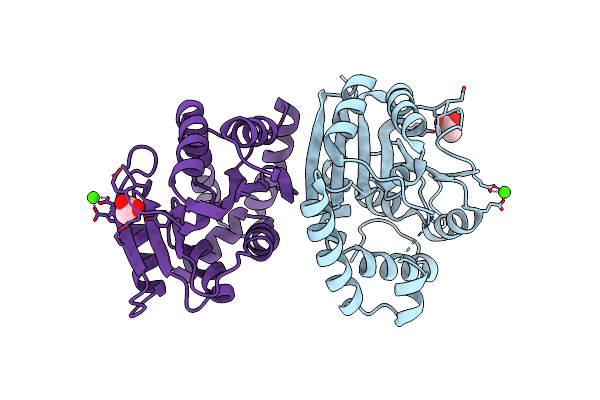
Crystal Structure Of Putative Oxidoreductase From Streptococcus Agalactiae 2603V/R
Organism: Streptococcus agalactiae serogroup v
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-10-21 Classification: OXIDOREDUCTASE |
 |
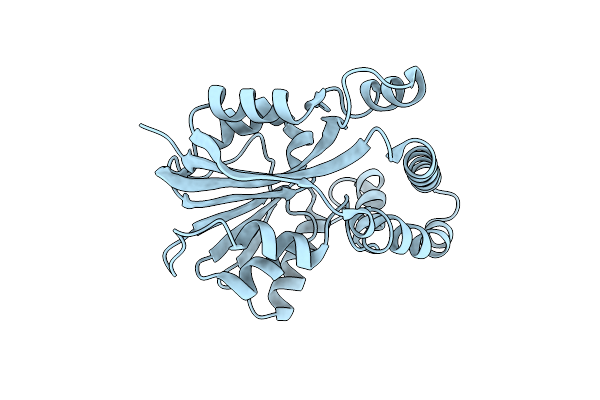
Crystal Structure Of Putative Methyltransferase-Mm_2633 From Methanosarcina Mazei .
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2008-09-09 Classification: structural genomics, unknown function Ligands: CA, ACT |
 |
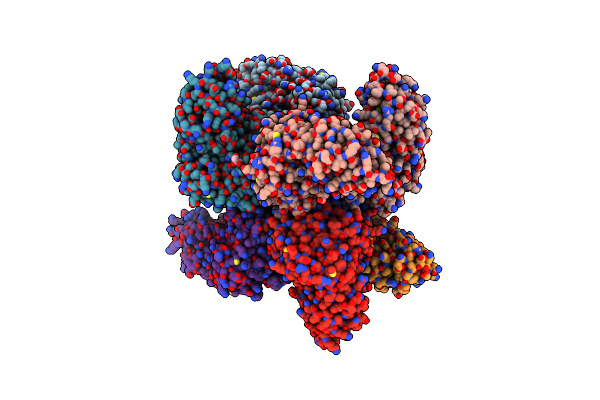
Crystal Structure Of Of Putative Methyltransferase From Bacteroides Vulgatus Atcc 8482
Organism: Bacteroides vulgatus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-09-02 Classification: TRANSFERASE |
 |
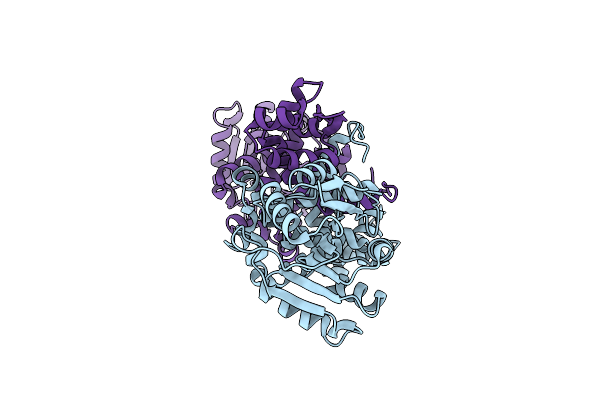
Crystal Structure Of Zn-Dependent Arginine Carboxypeptidase Complexed With Zinc
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2008-08-05 Classification: HYDROLASE Ligands: ZN, ARG, GOL |
 |
Crystal Structure Of Sam-Dependent Methyltransferase From Bacteroides Vulgatus Atcc 8482
Organism: Bacteroides vulgatus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2008-07-29 Classification: TRANSFERASE |
 |
Crystal Structure Of Putative Mandelate Racemase / Muconate Lactonizing Enzyme From Vibrionales Bacterium Swat-3
Organism: Vibrionales bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-07-01 Classification: ISOMERASE |
 |
Organism: Myxococcus xanthus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-05-06 Classification: ISOMERASE |
 |
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2008-03-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Organism: Chlamydophila abortus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-03-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: ZN |
 |
Crystal Structure Of Uncharacterized Protein Ism_01780 From Roseovarius Nubinhibens Ism
Organism: Roseovarius nubinhibens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2008-02-12 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: CA, NI |
 |
Crystal Structure Of An Uncharacterized Protein From Actinobacillus Succinogenes
Organism: Actinobacillus succinogenes
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2008-02-05 Classification: ISOMERASE Ligands: GOL, CL |
 |
Crystal Structure Of The N-Terminal Domain Of Response Regulator Receiver Protein From Methanoculleus Marisnigri Jr1
Organism: Methanoculleus marisnigri jr1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-02-05 Classification: SIGNALING PROTEIN Ligands: GOL |
 |
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2008-01-15 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: CL |
 |
Crystal Structure Of The C-Terminal Fragment Of Aaa+Atpase From Haemophilus Influenzae
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-01-01 Classification: NUCLEOTIDE BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Predicted Nucleotide-Binding Protein From Idiomarina Baltica Os145
Organism: Idiomarina baltica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-01-01 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: SO4, GOL |
 |
Crystal Structure Of Deoxyguanosinetriphosphate Triphosphohydrolase From Flavobacterium Sp. Med217
Organism: Leeuwenhoekiella blandensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2007-12-04 Classification: HYDROLASE |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ARG, GOL, MG |

