Search Count: 18,369
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: A1IZI |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2026-01-28 Classification: TRANSCRIPTION Ligands: A1IZ2 |
 |
Sam-Dependent C6-Fpp Methytransferase From Streptomyces Varsoviensis In Complex With Sah And Fpp
Organism: Streptomyces varsoviensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: FPP, SAH, PG4, PGE, PE8, PEG, EDO |
 |
Cryo-Em Structure Of Sars-Cov-2 Nsp10-Nsp14 (E191A) In Complex With T20P14-B Complex, Dimeric Form
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN/RNA Ligands: ZN, MG, EIF |
 |
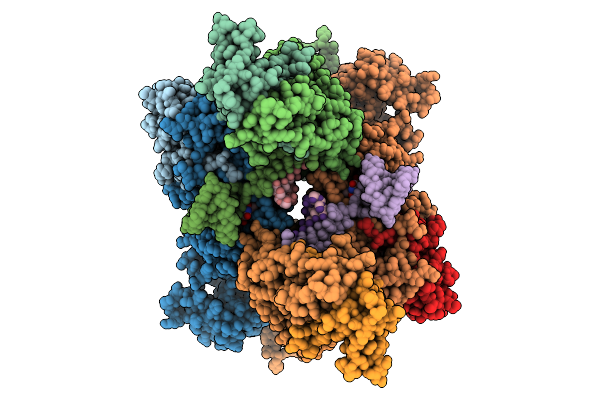
Cryo-Em Structure Of Sars-Cov-2 Nsp10-Nsp14 (E191A) In Complex With T20P14-B Complex, Protomer A Focused Refinement
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN/RNA Ligands: ZN, MG, EIF |
 |
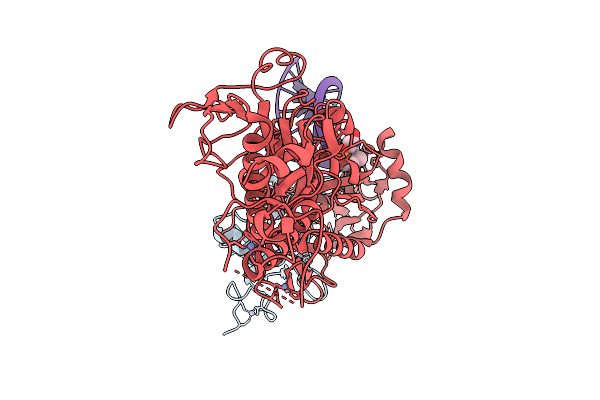
Cryo-Em Structure Of Sars-Cov-2 Nsp10-Nsp14 (E191A) In Complex With T20P14-S Complex, Tetrameric Form
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN/RNA Ligands: ZN, MG, K5X |
 |
Cryo-Em Structure Of Sars-Cov-2 Nsp10-Nsp14 (E191A) In Complex With T20P14-S Complex, Monomeric Form
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN/RNA Ligands: ZN, MG, K5X |
 |
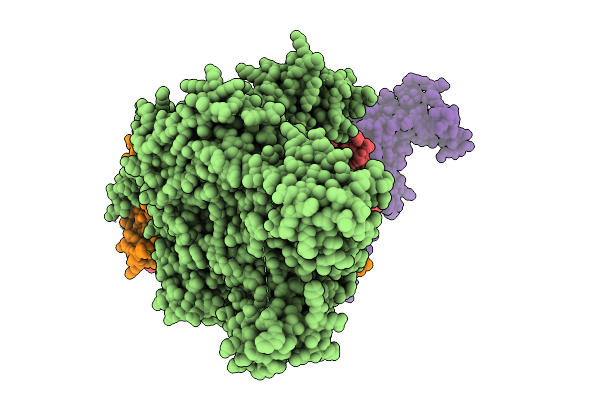
Cryo-Em Structure Of Dnmt 3A2/3B3 Tetramer Bound To A Di-Nucleosome With A 25 Base-Pair Linker
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, SAH |
 |
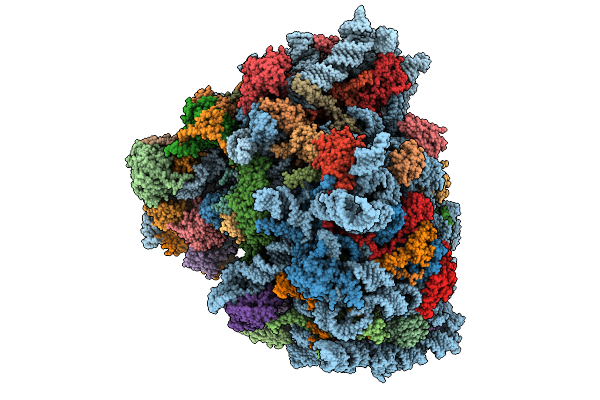
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.34 Å Release Date: 2026-01-21 Classification: CELL CYCLE |
 |
Rabbit 80S Ribosome In Complex With Erf1-Aaq, Stalled At The Stop Codon In Mutated F2A Sequence
Organism: Homo sapiens, Foot-and-mouth disease virus sat 2, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: RIBOSOME Ligands: MG, ZN, K, SPD, GTP, SF4 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, IMD, A1JKS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, IMD, EDO, CL, A1JMY |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: A1JMZ, IMD, EDO, CL, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: A1JM0, IMD, EDO, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, IMD, A1JM1 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: A1JM2, IMD, EDO, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2026-01-21 Classification: PROTEIN BINDING Ligands: NAD |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA Ligands: AN6 |
 |
Organism: Saccharomyces cerevisiae bmn1-35, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA Ligands: AN6 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA Ligands: AN6 |

