Search Count: 75
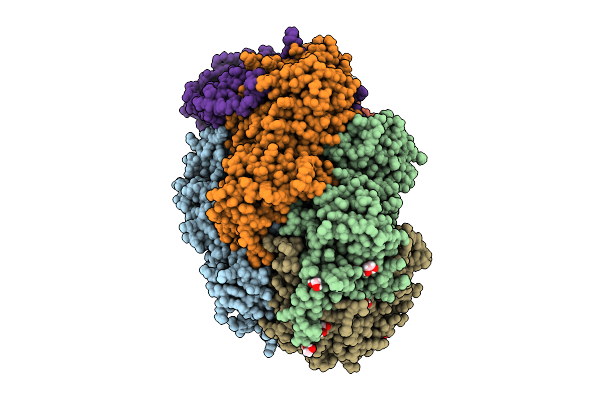 |
Methyl-Coenzyme M Reductase Of Anme-2D Candidatus Methanoperedens Vercelli Strain 1 From A Bioreactor Enrichment Culture
Organism: Candidatus methanoperedens sp.
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: K, F43, EDO, TP7, COM, SHT |
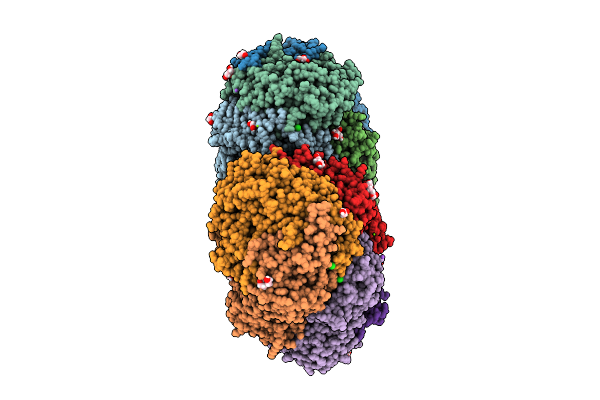 |
Krypton-Pressurized Methyl-Coenzyme M Reductase Of An Anme-2C Isolated From A Microbial Enrichment
Organism: Candidatus methanogasteraceae archaeon
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: KR, MG, PGE, EDO, CL, TP7, F43, PEG, GOL, COM, NA, K |
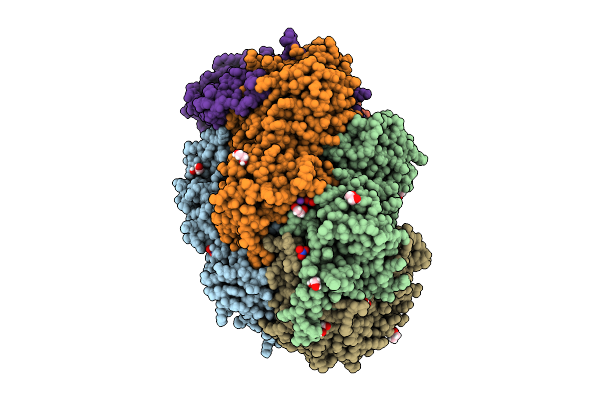 |
Methyl-Coenzyme M Reductase Of Anme-2D Candidatus Methanoperedens Sp. Blz2 From A Bioreactor Enrichment Culture
Organism: Candidatus methanoperedens sp. blz2
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2025-07-16 Classification: TRANSFERASE |
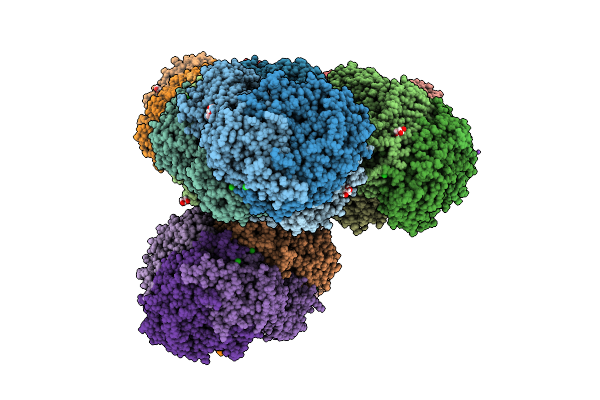 |
Organism: Candidatus methanogasteraceae archaeon
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: K, NA, CL, F43, COM, TP7, GOL, MPD, EDO |
 |
Cryoem Structure Of The Elp-Hdr Complex Of Methanothermobacter Marburgensis State 2 (Composite Structure)
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, 9S8 |
 |
Cryoem Structure Of The Elp-Hdr Complex Of Methanothermobacter Marburgensis State 2, Dimer (Composite Structure)
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, 9S8, FES |
 |
Cryoem Structure Of The Elp-Hdr Complex Of Methanothermobacter Marburgensis State 1 (Composite Structure)
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, 9S8 |
 |
Cryoem Structure Of The Hdr(Abc)2 Subunits Of The Elp-Hdr Complex Of Methanothermobacter Marburgensis
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: OXIDOREDUCTASE Ligands: 9S8, SF4, FAD |
 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: SHT, TP7, COM, F43, S5Q, ZN, ATP, MG |
 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, SHT, COM, TP7, S5Q |
 |
Methyl-Coenzyme M Reductase Activation Complex Binding To The A2 Component After Incubation With Atp
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, COM, TP7, SHT, S5Q, ZN, ATP, MG |
 |
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: MG, JCV, NA, COM |
 |
Cryo-Em Structure Of The Methanogenic Na+ Translocating N5-Methyl-H4Mpt:Com Methyltransferase Complex
Organism: Methanothermobacter marburgensis
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: MG, JCV, NA |
 |
Structure Of Mcrd (Methyl-Coenzyme M Reductase Operon Protein D) From Methanomassiliicoccus Luminyensis
Organism: Methanomassiliicoccus luminyensis b10
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-07-03 Classification: UNKNOWN FUNCTION Ligands: GOL |
 |
Mcrd Binds Asymmetrically To Methyl-Coenzyme M Reductase Improving Active Site Accessibility During Assembly
Organism: Methanosarcina acetivorans c2a
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: TRANSFERASE Ligands: COM, F43, TP7 |
 |
Organism: Methanosarcina acetivorans c2a
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: TRANSFERASE |
 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Hexameric, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, 9S8, ZN, MO, MGD |
 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Dimeric, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, 9S8, ZN, MO, MGD |
 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Heterodislfide Reductase Core And Mobile Arm In Conformational State 1, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, 9S8, FES |
 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Heterodisulfide Reductase Core And Mobile Arm In Conformational State 2, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, 9S8, FES |

