Search Count: 239
 |
Gtpbp1*Gcp*Phe-Trna*Ribosome In The Gtpase Activation-Like State, Structure Iii
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
 |
Gtpbp1*Gdp*Phe-Trna*Ribosome In The Post-Gtp Hydrolysis State, Structure Iv
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GDP, MG |
 |
Organism: Oryctolagus cuniculus, Saccharomyces cerevisiae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET |
 |
Organism: Oryctolagus cuniculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: SPD, ZN, ATP, MET |
 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, GCP, MG, K |
 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
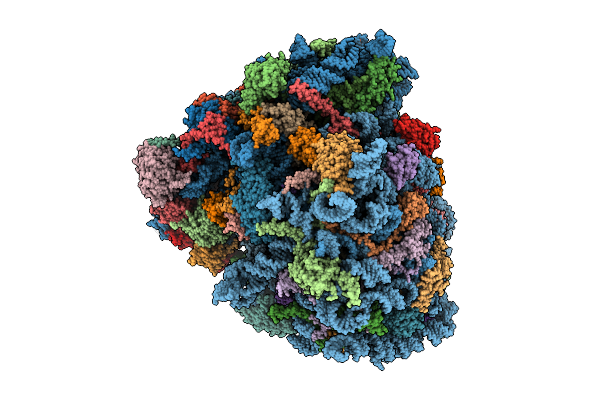 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, GCP, MG, K |
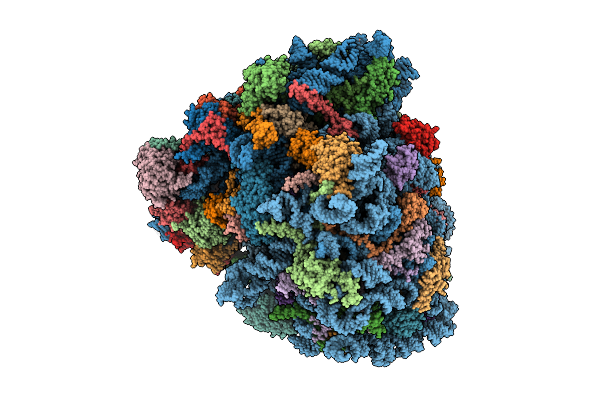 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
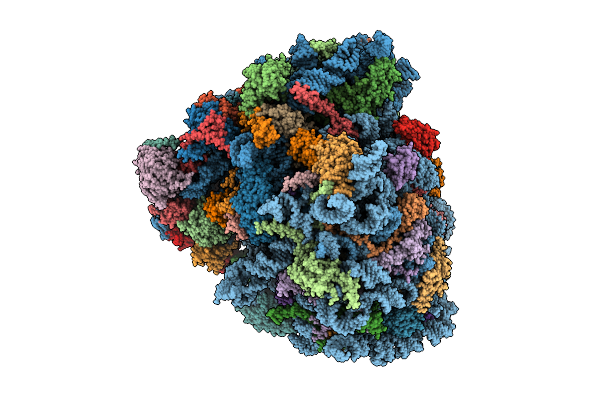 |
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, ATP, MET |
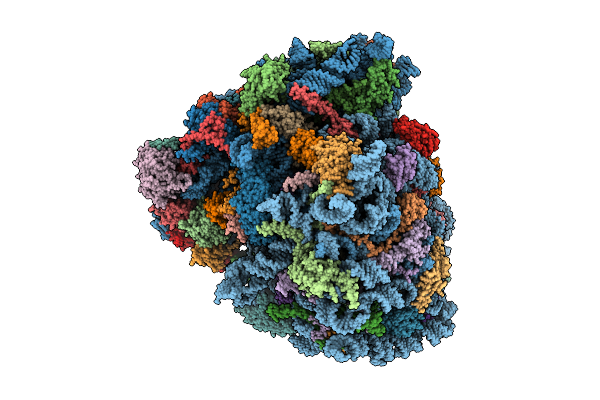 |
Organism: Oryctolagus cuniculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, ATP, MET |
 |
Organism: Burkholderia thailandensis
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: MET |
 |
Structure Of The Complex Between Human Lias And H-Protein In The Presence Of 5'-Deoxyadenosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: BIOSYNTHETIC PROTEIN Ligands: 5AD, SF4, F3S, PEG, PG4, MET, YVI, 2PE, PGE, PO4 |
 |
Structure Of Human Lias In The Presence Of 5'-Deoxyadenosine And Octanoyl-Modified Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, MET, 5AD, LYS, OCA |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Swivelled Conformation (Model Py48S-Auc-Swiv-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2.1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |

