Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
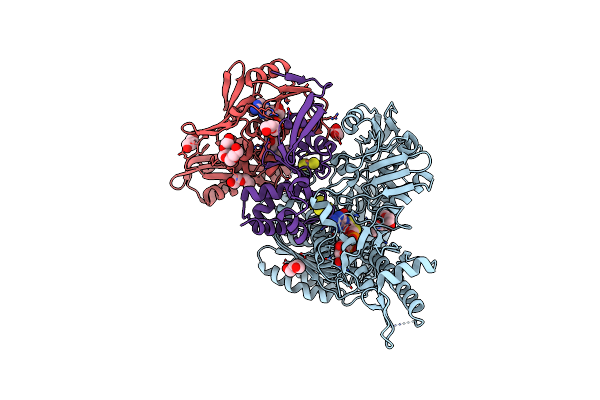
The Cryoem Structure Of The High Affinity Carbon Monoxide Dehydrogenase From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: CUN, MCN, FAD, FES |
 |
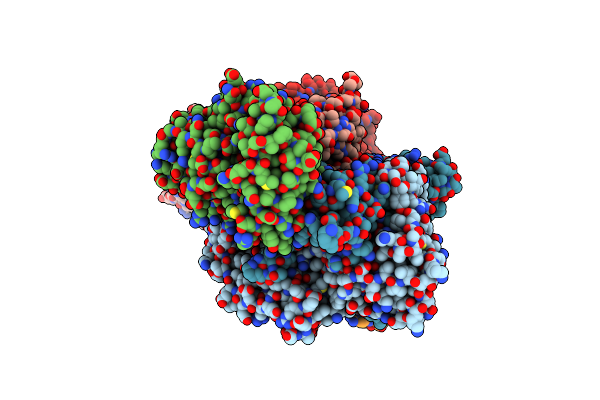
Organism: Paenarthrobacter nicotinovorans
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2022-01-26 Classification: OXIDOREDUCTASE Ligands: MCN, MO, FAD, FES |
 |
Organism: Methylobacillus sp. ky4400
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-01-24 Classification: OXIDOREDUCTASE Ligands: FES, SO4, FAD, SF4, GOL, MOS, MCN, IPA |
 |
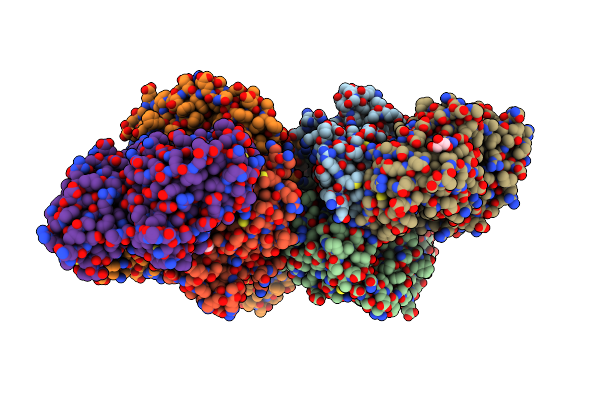
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE Ligands: FES, IOD, CL, ACT, GOL, SF4, FAD, MCN, MOS |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE Ligands: FES, IOD, ACT, CL, SF4, FAD, MCN, MOS, GOL |
 |
Organism: Sulfolobus tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-02-03 Classification: OXIDOREDUCTASE Ligands: MCN, MO, 1PE, PEG, FAD, PG4, ACY, FES |
 |
Organism: Eubacterium barkeri
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-06-30 Classification: OXIDOREDUCTASE Ligands: SE, NO3, MG, MOS, MCN, NIO, FAD, FES, CA |
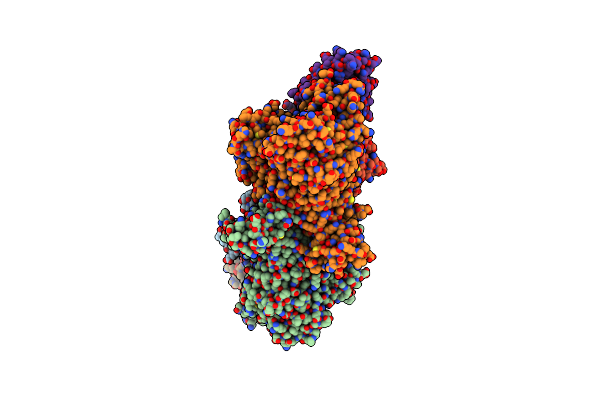 |
Organism: Oligotropha carboxidovorans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2005-10-11 Classification: OXIDOREDUCTASE Ligands: PO4, FES, CU, CUM, MCN, FAD |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-09-14 Classification: OXIDOREDUCTASE Ligands: FES, GOL, SO4, MCN, SMO, FAD |
 |
Organism: Oligotropha carboxidovorans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2002-12-23 Classification: OXIDOREDUCTASE Ligands: PO4, FES, CUM, MCN, FAD |
 |
Crystal Structure Of The Cu,Mo-Co Dehydrogenase (Codh); Cyanide-Inactivated Form
Organism: Oligotropha carboxidovorans
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2002-12-18 Classification: OXIDOREDUCTASE Ligands: PO4, FES, OMO, MCN, FAD |
 |
Crystal Structure Of The Cu,Mo-Co Dehydrogenase (Codh); Dithionite Reduced State
Organism: Oligotropha carboxidovorans
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2002-12-18 Classification: OXIDOREDUCTASE Ligands: PO4, FES, CUN, MCN, FAD |
 |
Crystal Structure Of The Mo,Cu-Co Dehydrogenase (Codh), N-Butylisocyanide-Bound State
Organism: Oligotropha carboxidovorans
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2002-12-18 Classification: OXIDOREDUCTASE Ligands: PO4, FES, CUB, MCN, FAD |
 |
Crystal Structure Of The Cu,Mo-Co Dehydrogenase (Codh); Carbon Monoxide Reduced State
Organism: Oligotropha carboxidovorans
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2002-12-18 Classification: OXIDOREDUCTASE Ligands: PO4, FES, CUN, MCN, FAD |
 |
Crystal Structure Of The Aldehyde Oxidoreductase From Desulfovibrio Desulfuricans Atcc 27774
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2000-03-22 Classification: OXIDOREDUCTASE Ligands: FES, 2MO, MCN |