Search Count: 40
 |
Acla From Tenacibaculum Discolor In Complex With The C8-N-Acyl Cyclolysine Reaction Product (C8-Acl)
Organism: Tenacibaculum discolor
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: A1CA3, MAE, PEG |
 |
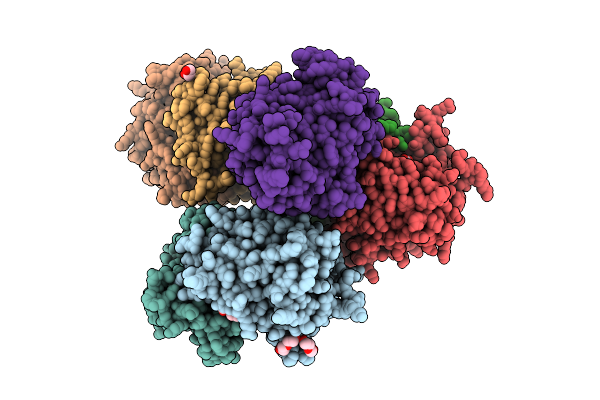
Crystal Structure Of Human Glucose-6-Phosphate Isomerase With Maleate Ligand
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-03-12 Classification: ISOMERASE Ligands: TRS, PG4, MAE, PGE |
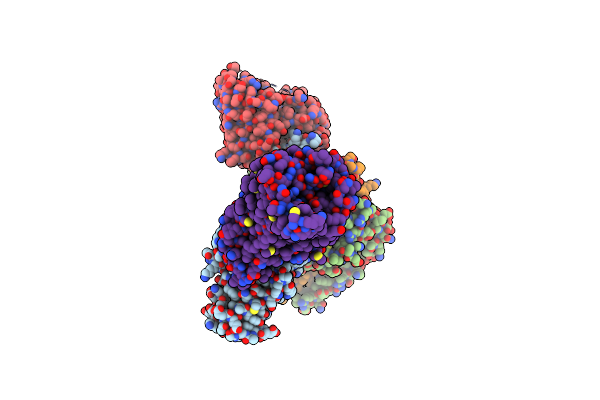 |
Organism: Homo sapiens, Rattus norvegicus, Synthetic construct, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: MEMBRANTE PROTEIN/IMMUNE SYSTEM Ligands: MAE |
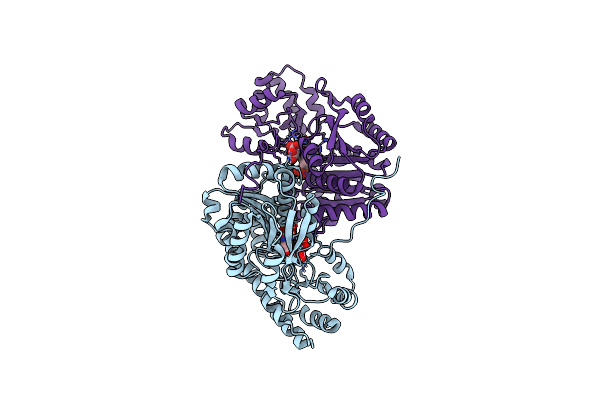 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Aifs Bound To Maleic Acid At 100 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: MAE, PLP |
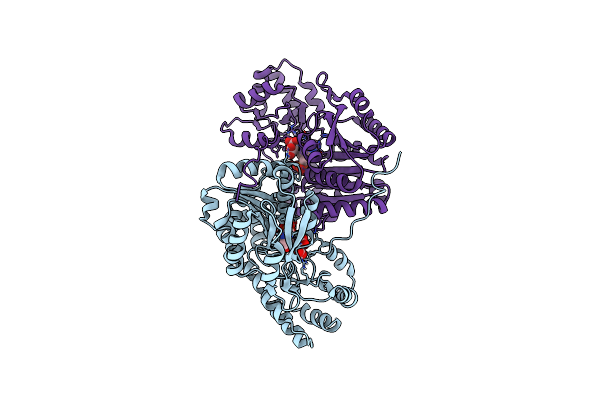 |
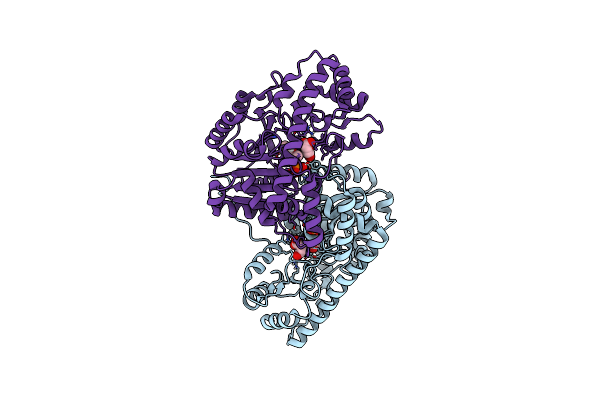
Crystal Structure Of Wild-Type E. Coli Aspartate Aminotransferase Bound To Maleate At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
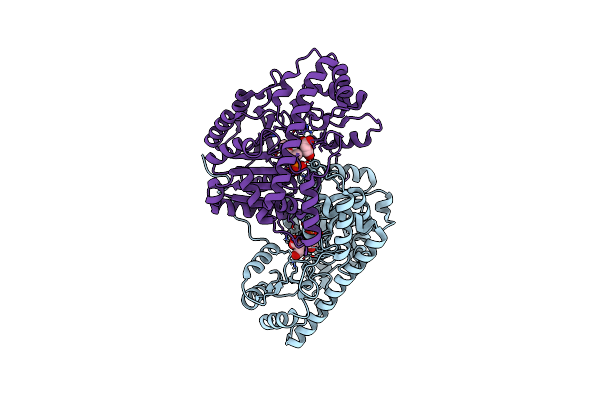
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfit Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
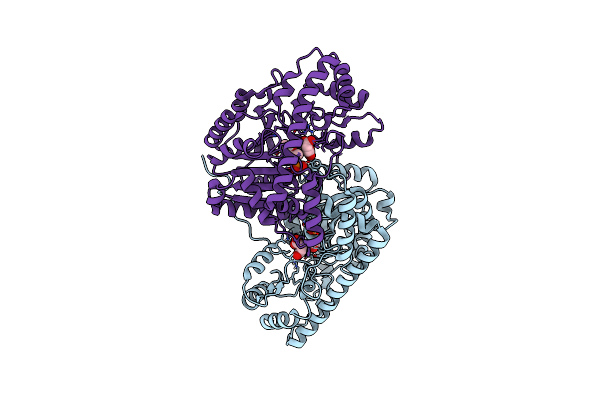
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfiy Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
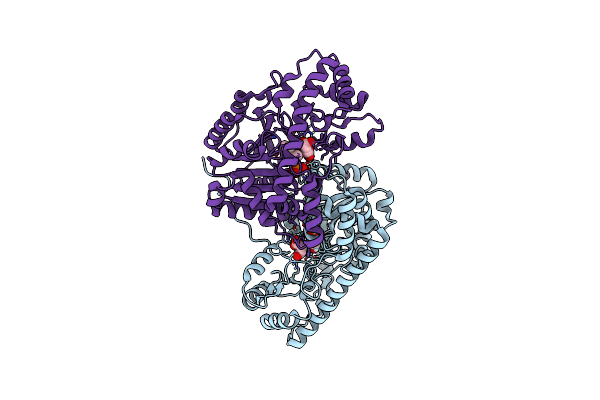
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Hex Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfcs Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of Pyrococcus Horikoshii Quinolinate Synthase (Nada) With Bound Maleate And Fe4S4 Cluster
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2016-07-27 Classification: TRANSFERASE Ligands: SF4, MAE, CL, NH4 |
 |
Crystal Structure Of The Maleamate Amidase Ami(C149A) In Complex With Maleate From Pseudomonas Putida S16
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2014-07-16 Classification: HYDROLASE Ligands: MAE |
 |
Crystal Structure Of The Maleate Isomerase Iso(C200A) From Pseudomonas Putida S16 With Maleate
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-07-24 Classification: ISOMERASE Ligands: MAE |
 |
Crystal Structure Of An Enolase Family Member From Vibrio Harveyi (Efi-Target 501692) With Homology To Mannonate Dehydratase, With Mg, Glycerol And Dicarboxylates Bound (Mixed Loops, Space Group I4122)
Organism: Vibrio harveyi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-08-29 Classification: LYASE Ligands: CL, GOL, MLA, MG, MAE |
 |
Crystal Structure Of The Aminopeptidase N Family Protein Q5Qty1 From Idiomarina Loihiensis. Northeast Structural Genomics Consortium Target Ilr60.
Organism: Idiomarina loihiensis l2tr
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2012-08-01 Classification: HYDROLASE Ligands: MAE, ZN |
 |
Organism: Moorella thermoacetica
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2011-12-14 Classification: LIGASE Ligands: SO4, AGS, TOE, MAE |
 |
E. Coli Aspartate Transcarbamoylase Complexed With N-Phosphonacetyl-L-Asparagine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-08-28 Classification: TRANSFERASE Ligands: 1IP, MPD, ZN, MAE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2006-11-14 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2006-11-14 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Maleic Acid-Bound Structure Of Srhept Mutant Of E. Coli Aspartate Aminotransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2004-10-05 Classification: TRANSFERASE Ligands: MAE |
 |
Crystal Structure Of Trypanosoma Cruzi Trypanothione Reductase In Complex With The Inhibitor Quinacrine Mustard
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-05-06 Classification: OXIDOREDUCTASE Ligands: FAD, MAE, QUM |

