Search Count: 15
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: TRANSPORT PROTEIN |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: TRANSPORT PROTEIN |
 |
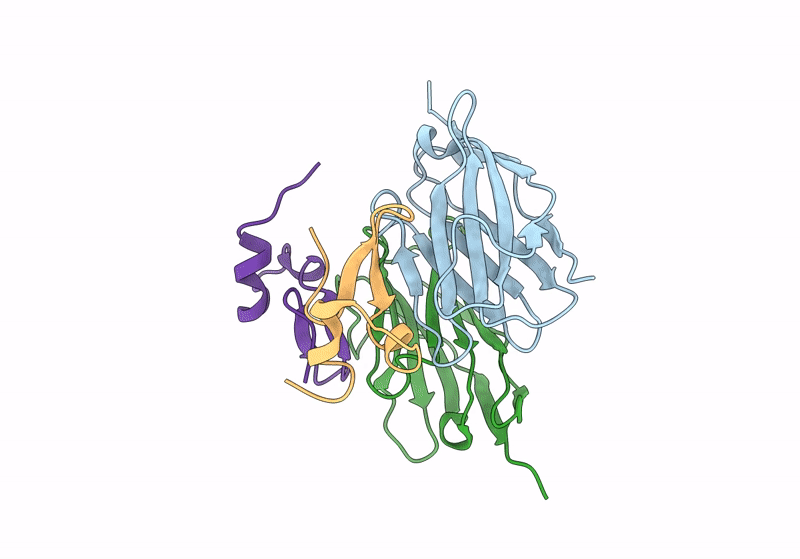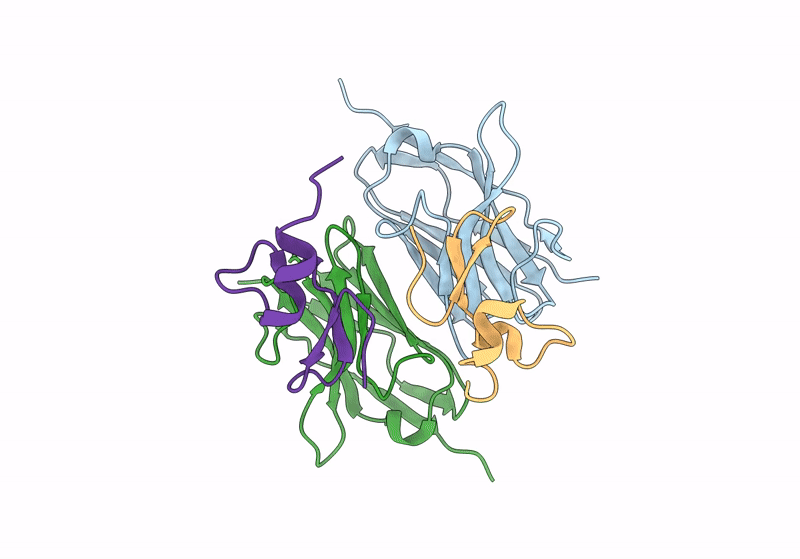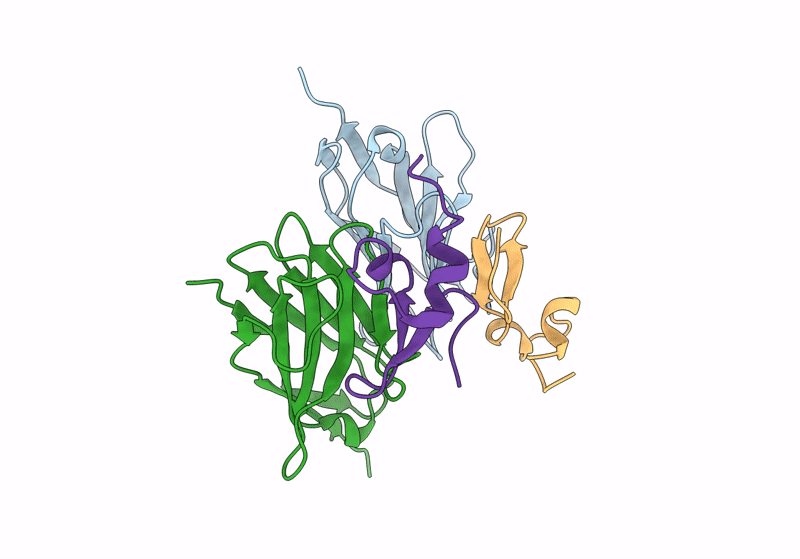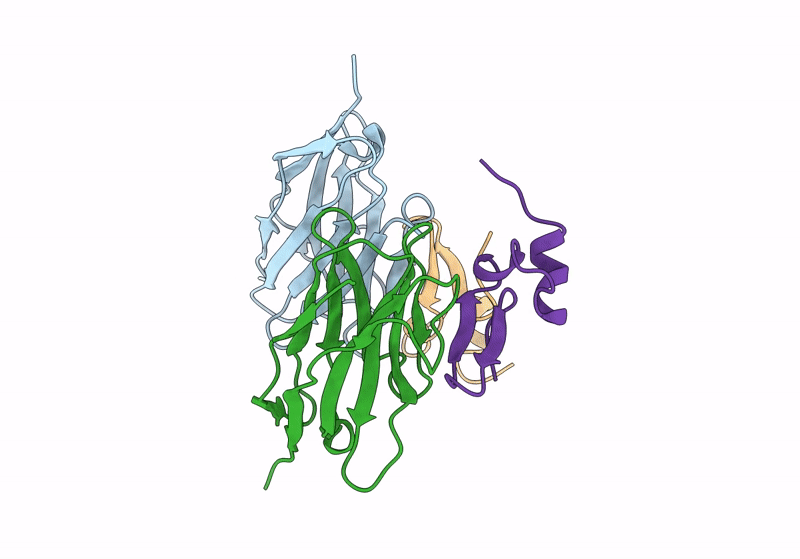
Organism: Vicugna pacos, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-01-03 Classification: IMMUNE SYSTEM |
 |
Organism: Vicugna pacos, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-01-03 Classification: IMMUNE SYSTEM |
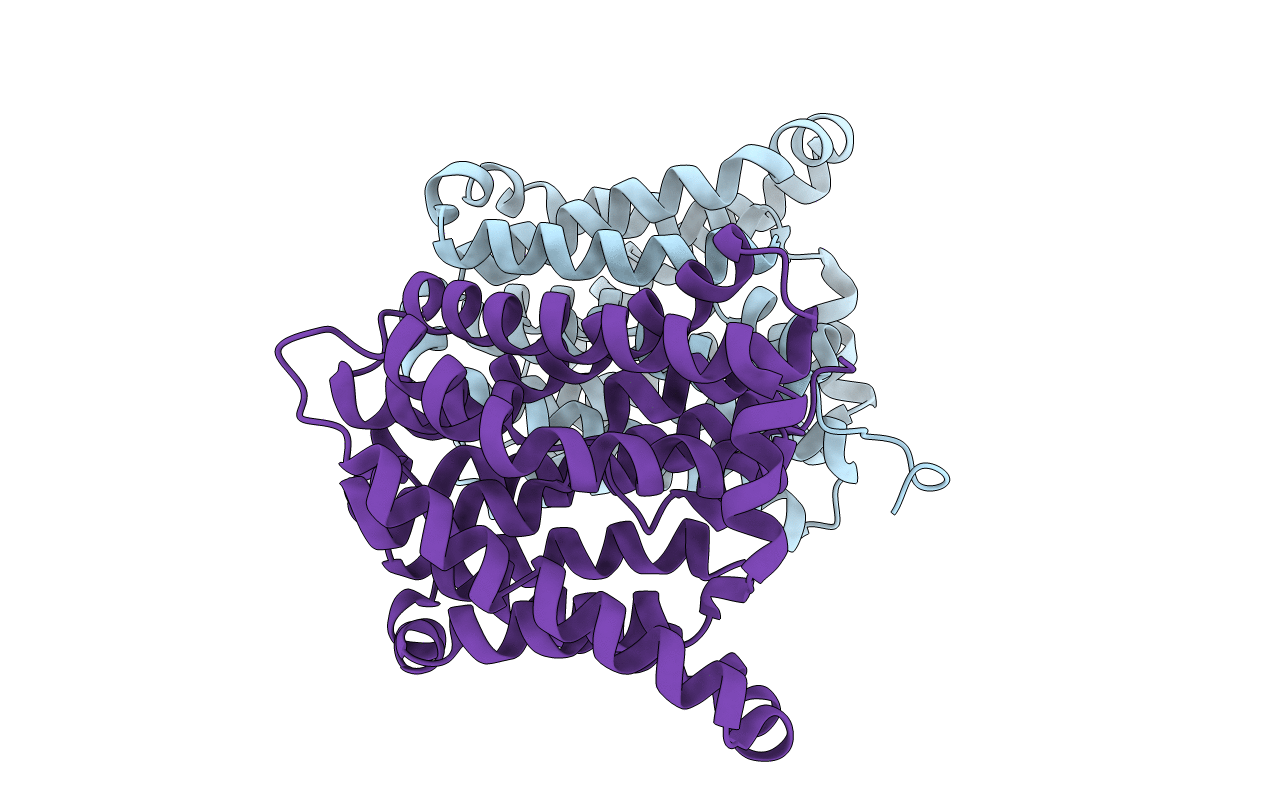 |
Crystal Structure Of A Cysteine-Pair Mutant (Y113C-P190C) Of A Bacterial Bile Acid Transporter Before Disulfide Bond Formation
Organism: Yersinia frederiksenii
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-01-13 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of A Second Cysteine-Pair Mutant (V110C-I197C) Of A Bacterial Bile Acid Transporter Before Disulfide Bond Formation
Organism: Yersinia frederiksenii
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-01-13 Classification: TRANSPORT PROTEIN Ligands: HG |
 |
Crystal Structure Of A Cysteine-Pair Mutant (P10C-S291C) Of A Bacterial Bile Acid Transporter In An Inward-Facing State Complexed With Citrate
Organism: Yersinia frederiksenii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: CIT, A6L |
 |
Crystal Structure Of A Cysteine-Pair Mutant (P10C-S291C) Of A Bacterial Bile Acid Transporter In An Inward-Facing State Complexed With Glycine And Sodium
Organism: Yersinia frederiksenii
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: GLY, NA, 2PE, A6L |
 |
Crystal Structure Of A Cysteine-Pair Mutant (P10C-S291C) Of A Bacterial Bile Acid Transporter In An Inward-Facing State Complexed With Sulfate
Organism: Yersinia frederiksenii
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: A6L, SO4 |
 |
Crystal Structure Of A Cysteine-Pair Mutant (P10C-S291C) Of A Bacterial Bile Acid Transporter In An Inward-Facing Apo-State
Organism: Yersinia frederiksenii
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: A6L |
 |
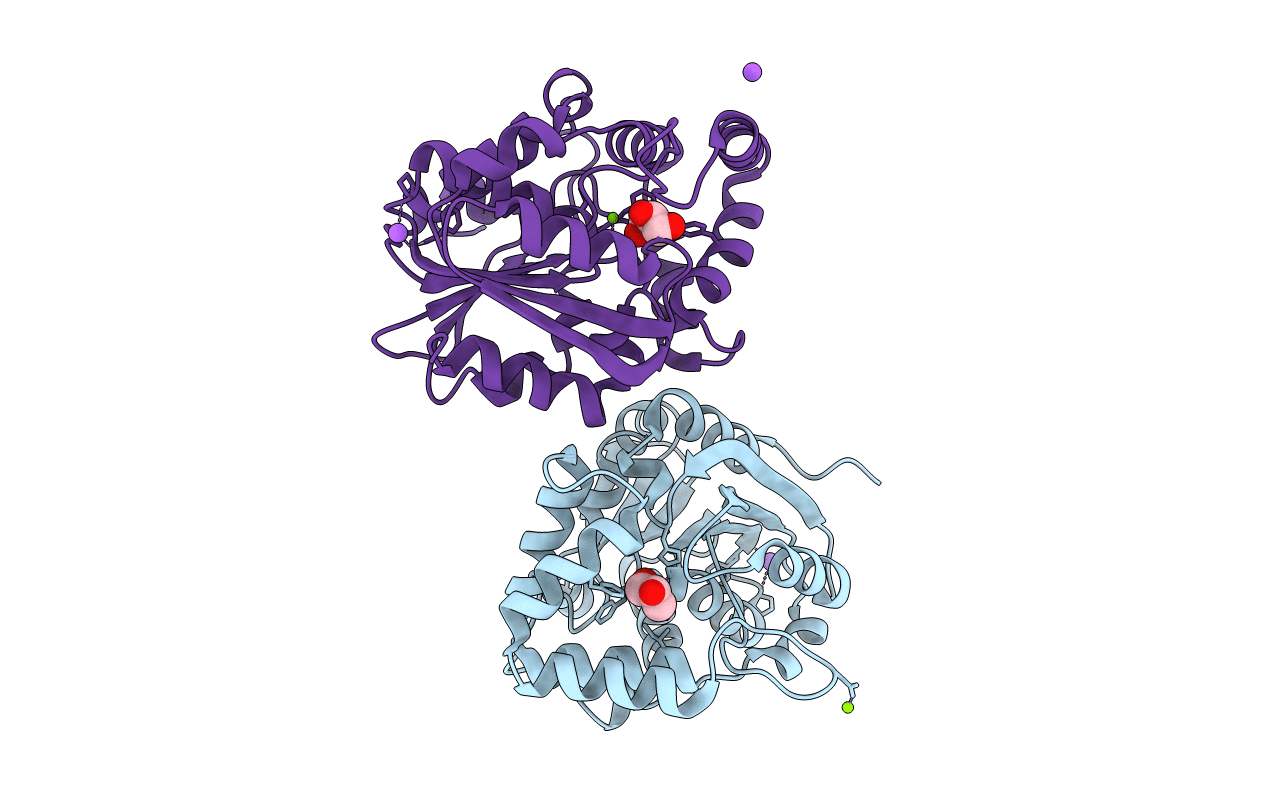
Crystal Structure Of A Cysteine-Pair Mutant (Y113C-P190C) Of A Bacterial Bile Acid Transporter Trapped In An Outward-Facing Conformation
Organism: Yersinia frederiksenii
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2020-12-09 Classification: TRANSPORT PROTEIN Ligands: CIT, A6L |
 |
Organism: Striga hermonthica
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: MG, NA, GOL |
 |
Organism: Striga hermonthica
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: EDO, MG |
 |
Organism: Striga hermonthica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: DIO, MG, EDO |
 |
Organism: Striga hermonthica
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: PEG, GOL, EDO |

