Search Count: 24
 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
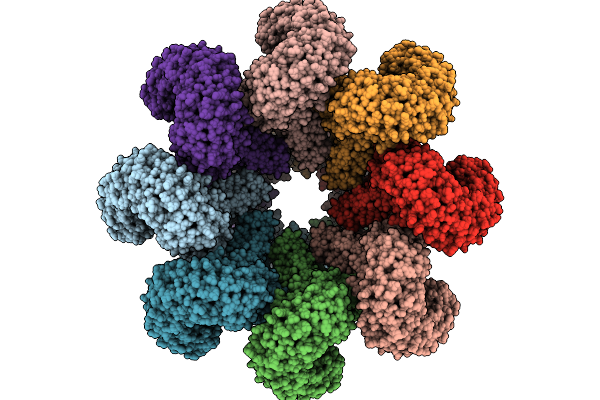
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat (N-To-N Arrangement)
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
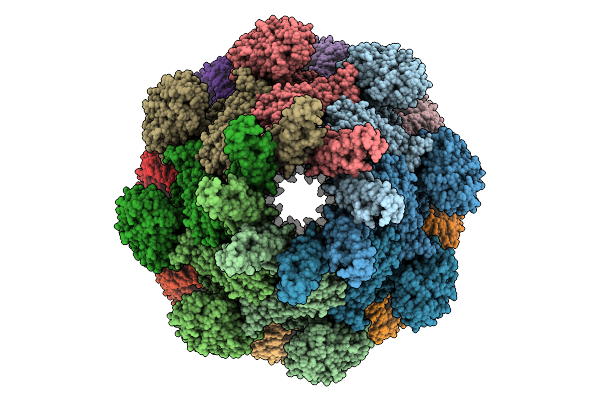 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat In 'Back-To-Back' Arrangement
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
Organism: Homo sapiens, Unidentified
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-05-07 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.74 Å Release Date: 2025-05-07 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.18 Å Release Date: 2025-05-07 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1BF3 |
 |
Organism: Schizosaccharomyces pombe 972h-, Streptococcus sp. group g
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: CELL CYCLE |
 |
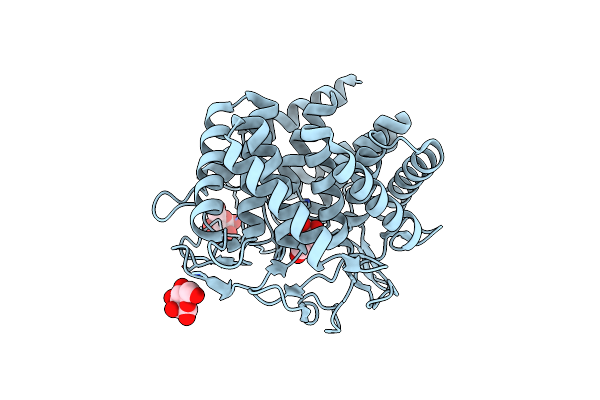
Crystal Structure Of Bromodomain Of Human Cbp In Complex With The Inhibitor Czl-046
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2024-10-16 Classification: PROTEIN BINDING Ligands: A1L3S, CL |
 |
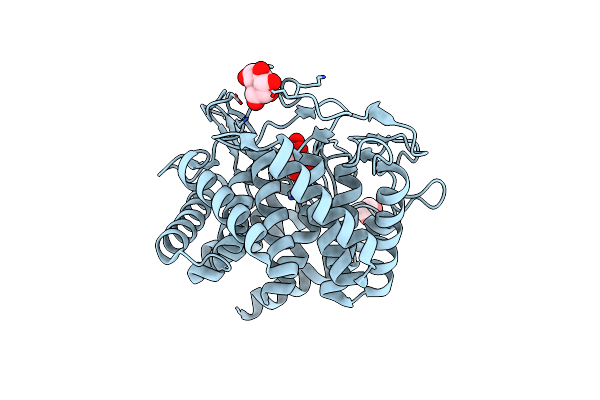
The Crystal Structure Of Circular Mannose With Mutant H247F Of The Cellobiose 2-Epimerase From Caldicellulosiruptor Saccharolyticus
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-25 Classification: ISOMERASE Ligands: BMA |
 |
The Crystal Structure Of Linear Mannose With Mutant H247F Of The Cellobiose 2-Epimerase From Caldicellulosiruptor Saccharolyticus
Organism: Caldicellulosiruptor saccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-10-25 Classification: ISOMERASE Ligands: DNO, BMA, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-01-25 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Csce With Ligand To Have A Insight Into The Catalytic Mechanism
Organism: Caldicellulosiruptor saccharolyticus dsm 8903
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-12-01 Classification: PROTEIN BINDING Ligands: CL |
 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-03-31 Classification: LIGASE Ligands: TSQ |
 |
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-03-31 Classification: LIGASE |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-09-09 Classification: FLUORESCENT PROTEIN Ligands: GOL, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2019-09-18 Classification: ANTITUMOR PROTEIN Ligands: CQF |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-09-18 Classification: ANTITUMOR PROTEIN Ligands: CQF, MES |
 |
Organism: Caldibacillus thermoamylovorans
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-06-19 Classification: ISOMERASE |
 |
The Structure Of Cellobiose 2-Epimerase From Spirochaeta Thermophila Dsm 6192
Organism: Spirochaeta thermophila dsm 6192
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-04-10 Classification: ISOMERASE Ligands: EDO |
 |
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: FAD, 1W6 |

