Search Count: 52
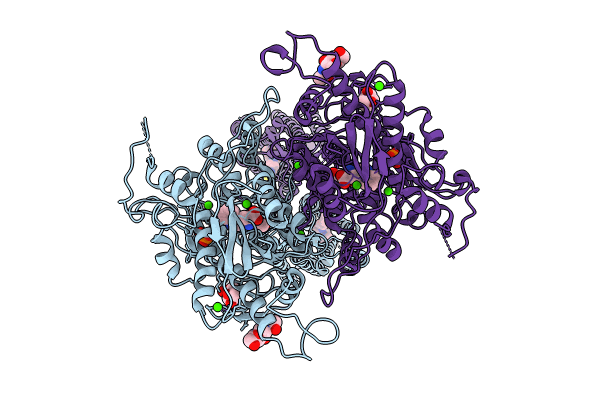 |
Structure Of Calcium-Sensing Receptor In Complex With Positive Allosteric Modulator '6218
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: NAG, CA, TRP, PO4, A1ATP |
 |
Structure Of Calcium-Sensing Receptor In Complex With Positive Allosteric Modulator '54149
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: NAG, CA, TRP, PO4, A1ATX |
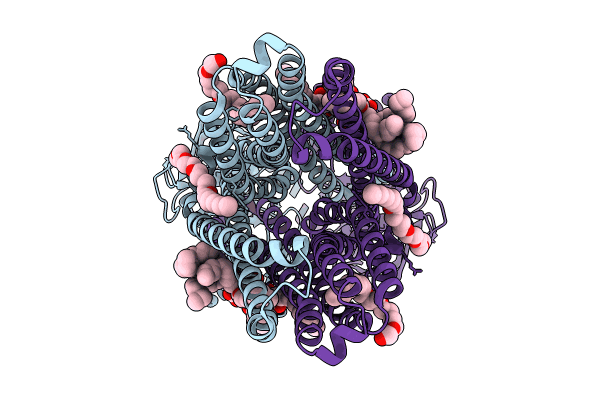 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN Ligands: CXE, KDL |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN |
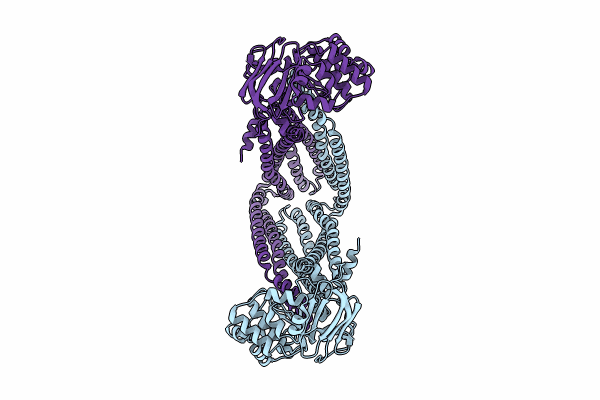 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TRANSPORT PROTEIN |
 |
5-Ht2Ar Bound To Lisuride In Complex With A Mini-Gq Protein And An Active-State Stabilizing Single-Chain Variable Fragment (Scfv16) Obtained By Cryo-Electron Microscopy (Cryoem)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: MEMBRANE PROTEIN Ligands: H8G |
 |
5Ht2Ar-Minigq Heterotrimer In Complex With A Novel Agonist Obtained From Large Scale Docking
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: MEMBRANE PROTEIN Ligands: YEQ |
 |
The Crystal Structure Of Dgtpalphase-Sp:Dnapre-Ii:Pol X Substrate Ternary Complex
Organism: African swine fever virus (strain badajoz 1971 vero-adapted), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-01-17 Classification: REPLICATION/DNA Ligands: MN, 7Q6 |
 |
Organism: African swine fever virus (strain badajoz 1971 vero-adapted), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-01-17 Classification: REPLICATION/DNA Ligands: MN, FMT |
 |
The Crystal Structure Of Native Dgtp:Dnapre-I:Pol X Substrate Ternary Complex
Organism: African swine fever virus (strain badajoz 1971 vero-adapted), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-01-10 Classification: REPLICATION/DNA Ligands: DGT, MN, SO4 |
 |
The Crystal Structure Of Dgtpalphase-Rp:Dnapre-Ii:Pol X Substrate Ternary Complex
Organism: African swine fever virus (strain badajoz 1971 vero-adapted), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-01-10 Classification: REPLICATION/DNA Ligands: MN, 7Q6 |
 |
Organism: African swine fever virus (strain badajoz 1971 vero-adapted), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-10 Classification: REPLICATION/DNA Ligands: MG, SO4, 8EB, GMS |
 |
Organism: African swine fever virus (strain badajoz 1971 vero-adapted), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-10 Classification: REPLICATION/DNA Ligands: MG, SO4 |
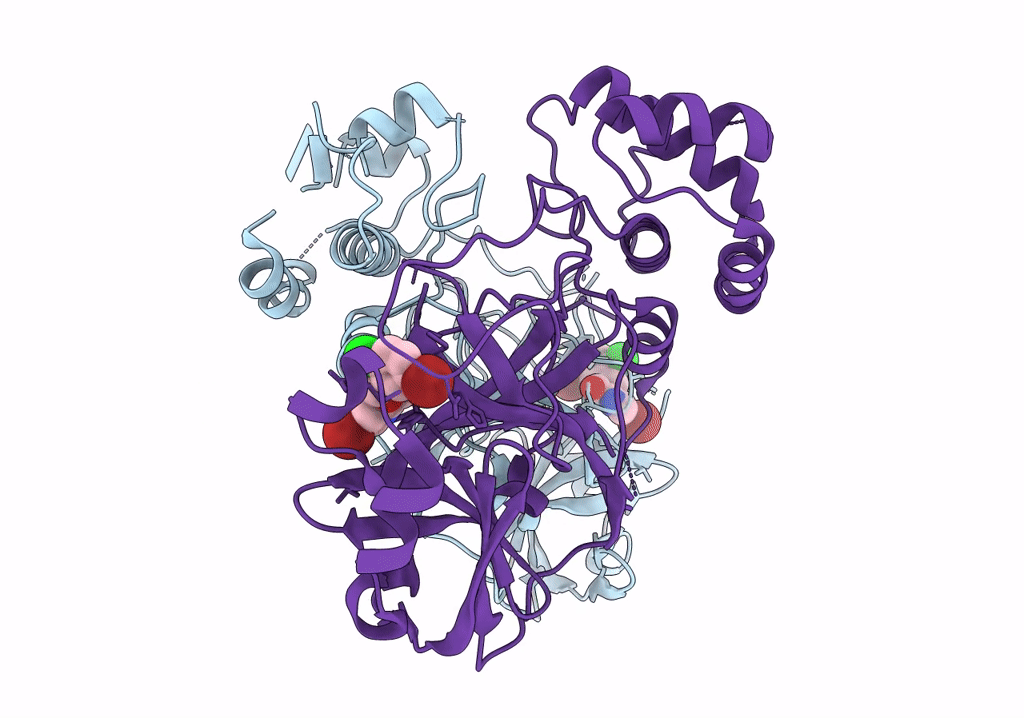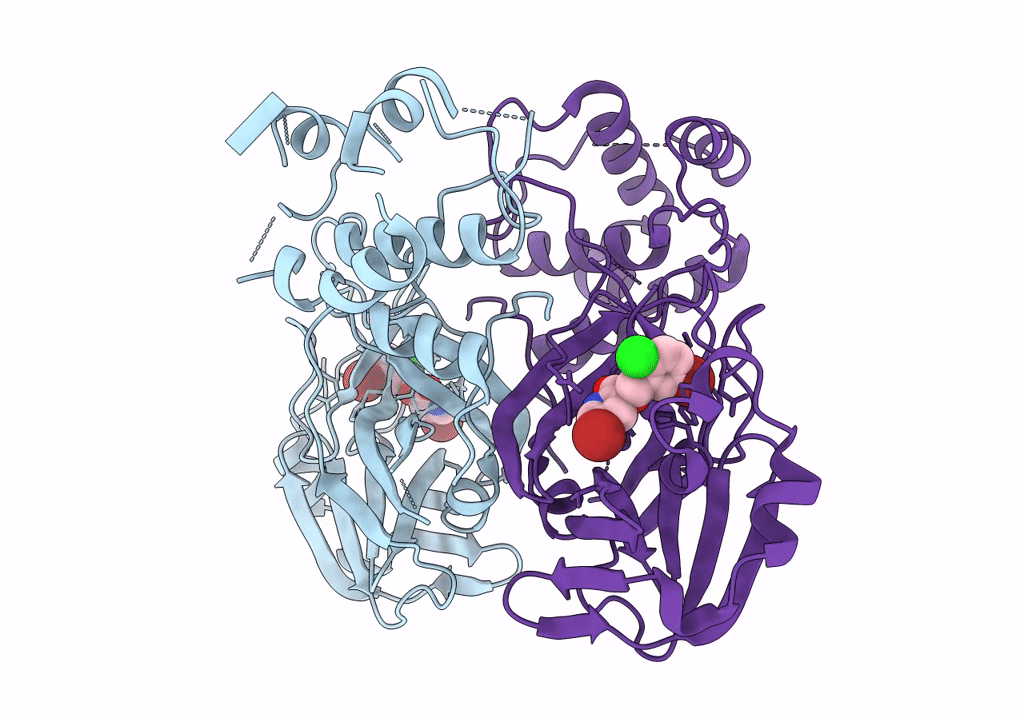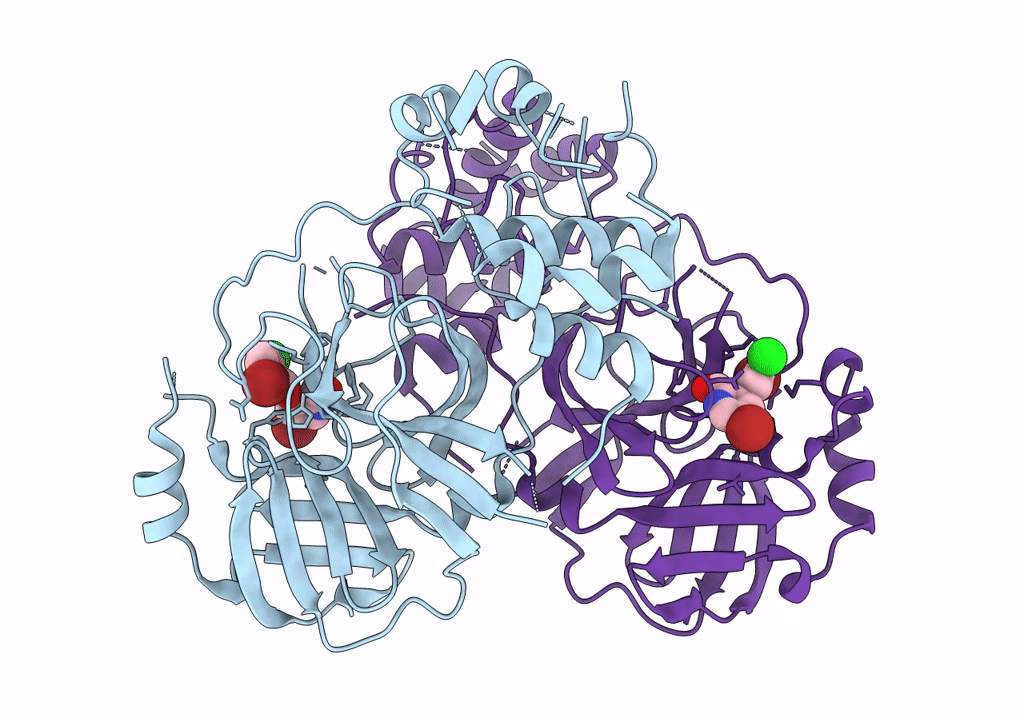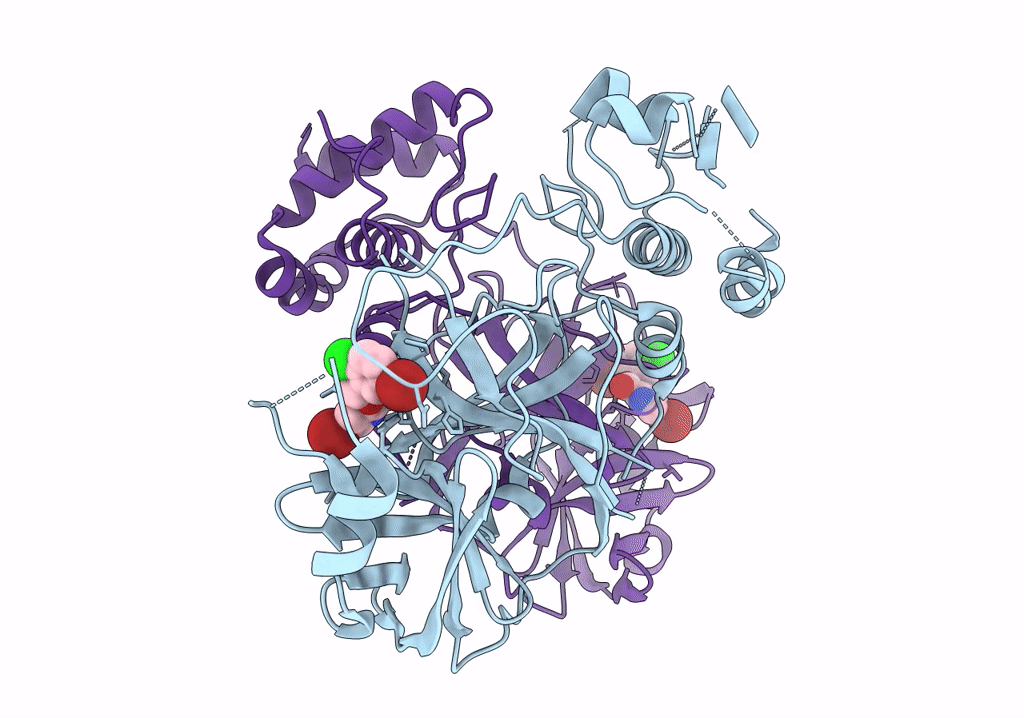 |
Virtual Screening For Novel Sars-Cov-2 Main Protease Non-Covalent And Covalent Inhibitors
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-07-05 Classification: VIRAL PROTEIN,HYDROLASE/INHIBITOR Ligands: U0R |
 |
Virtual Screening For Novel Sars-Cov-2 Main Protease Non-Covalent And Covalent Inhibitors
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-06-28 Classification: VIRAL PROTEIN,HYDROLASE/INHIBITOR Ligands: TKX |
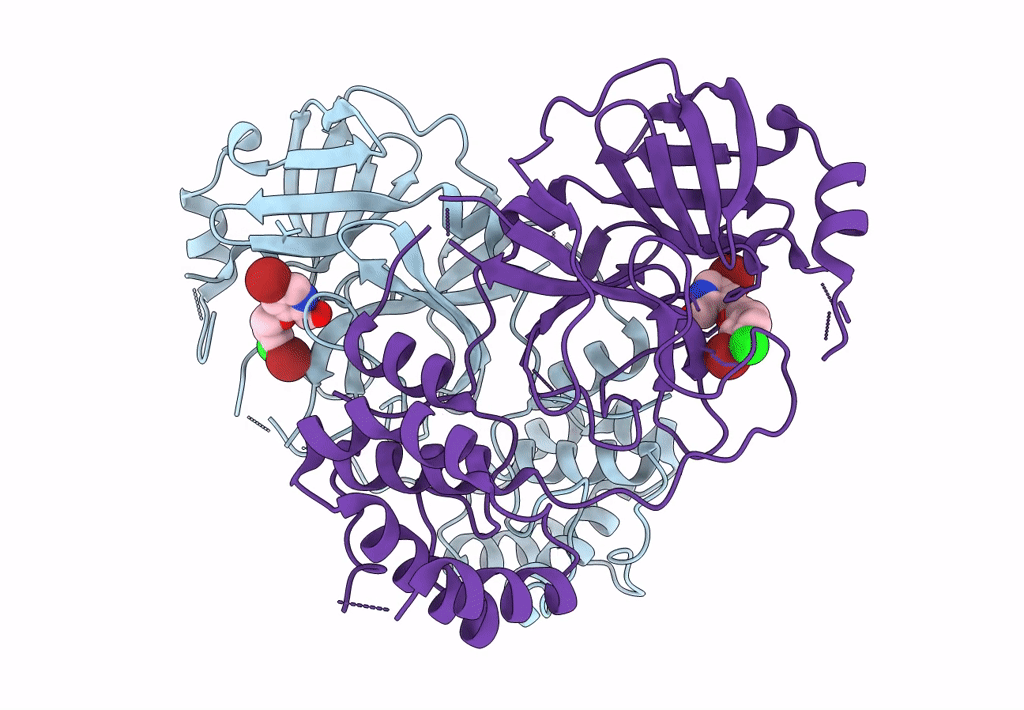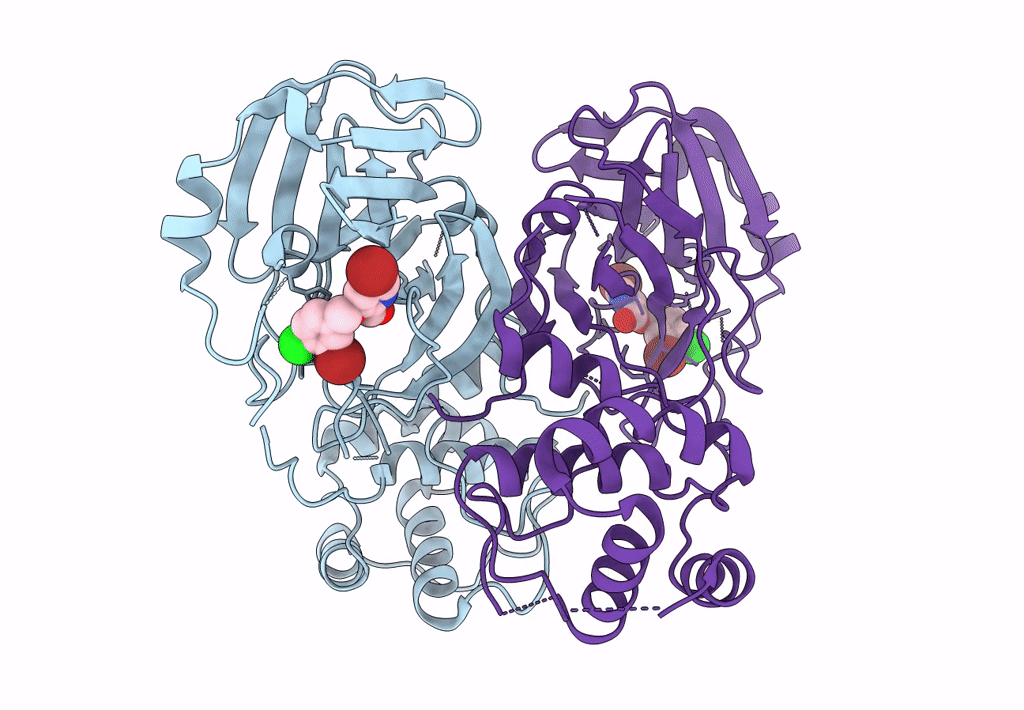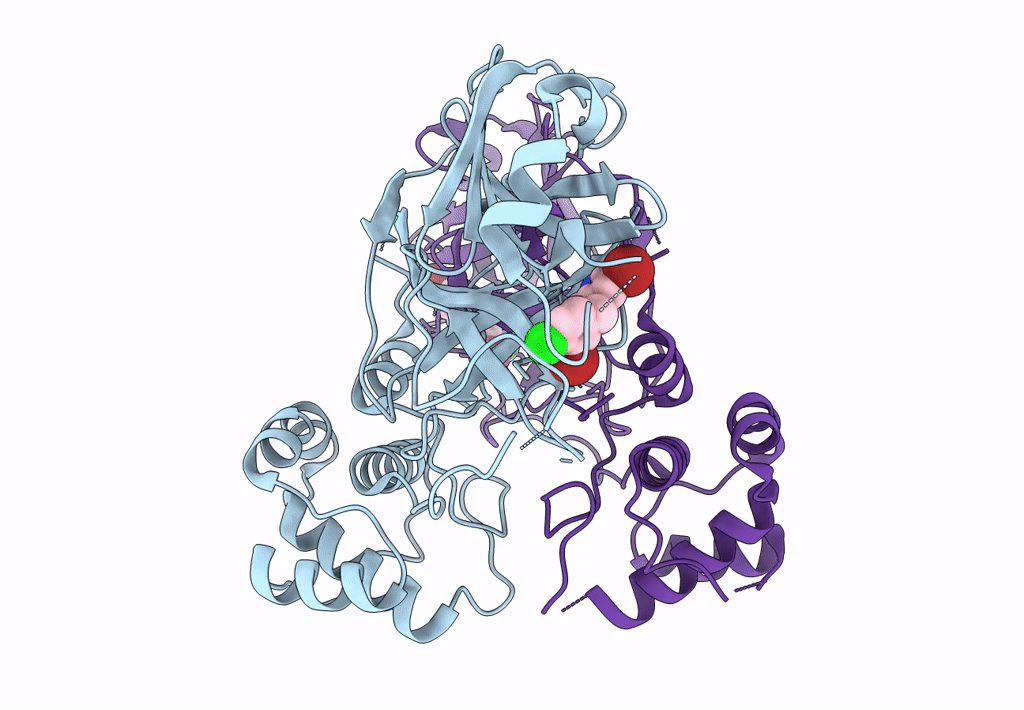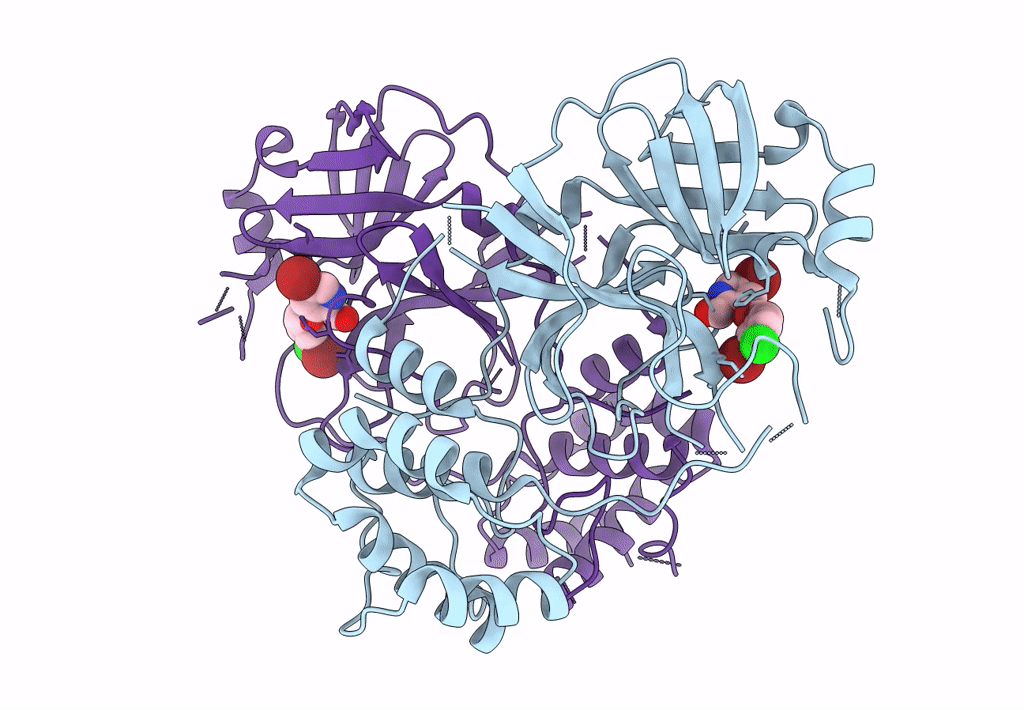 |
Virtual Screening For Novel Sars-Cov-2 Main Protease Non-Covalent And Covalent Inhibitors
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-06-28 Classification: VIRAL PROTEIN,HYDROLASE/INHIBITOR Ligands: TNI |
 |
Virtual Screening For Novel Sars-Cov-2 Main Protease Non-Covalent And Covalent Inhibitors
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-28 Classification: VIRAL PROTEIN,HYDROLASE/INHIBITOR Ligands: U1J |
 |
Virtual Screening For Novel Sars-Cov-2 Main Protease Non-Covalent And Covalent Inhibitors
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-06-28 Classification: VIRAL PROTEIN,HYDROLASE/INHIBITOR Ligands: U1R |

