Search Count: 6
 |
Ftse Structure From Streptococcus Pneumoniae In Complex With Adp (Space Group P 1)
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2020-09-02 Classification: CELL CYCLE Ligands: ADP |
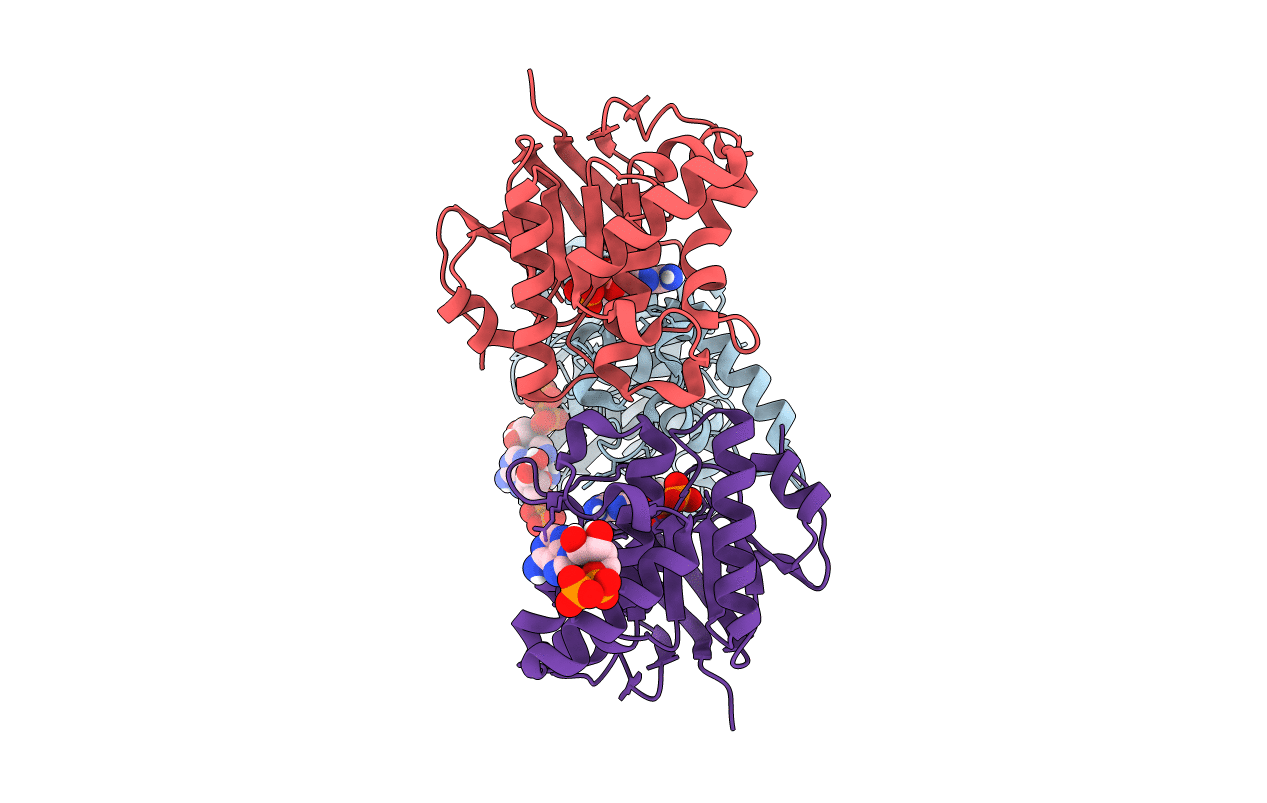 |
Ftse Structure From Streptococus Pneumoniae In Complex With Adp At 1.57 A Resolution (Spacegroup P 21)
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-09-02 Classification: CELL CYCLE Ligands: ADP |
 |
Ftse Structure Of Streptococcus Pneumoniae In Complex With Amppnp At 2.4 A Resolution
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-09-02 Classification: CELL CYCLE Ligands: ADP, ANP |
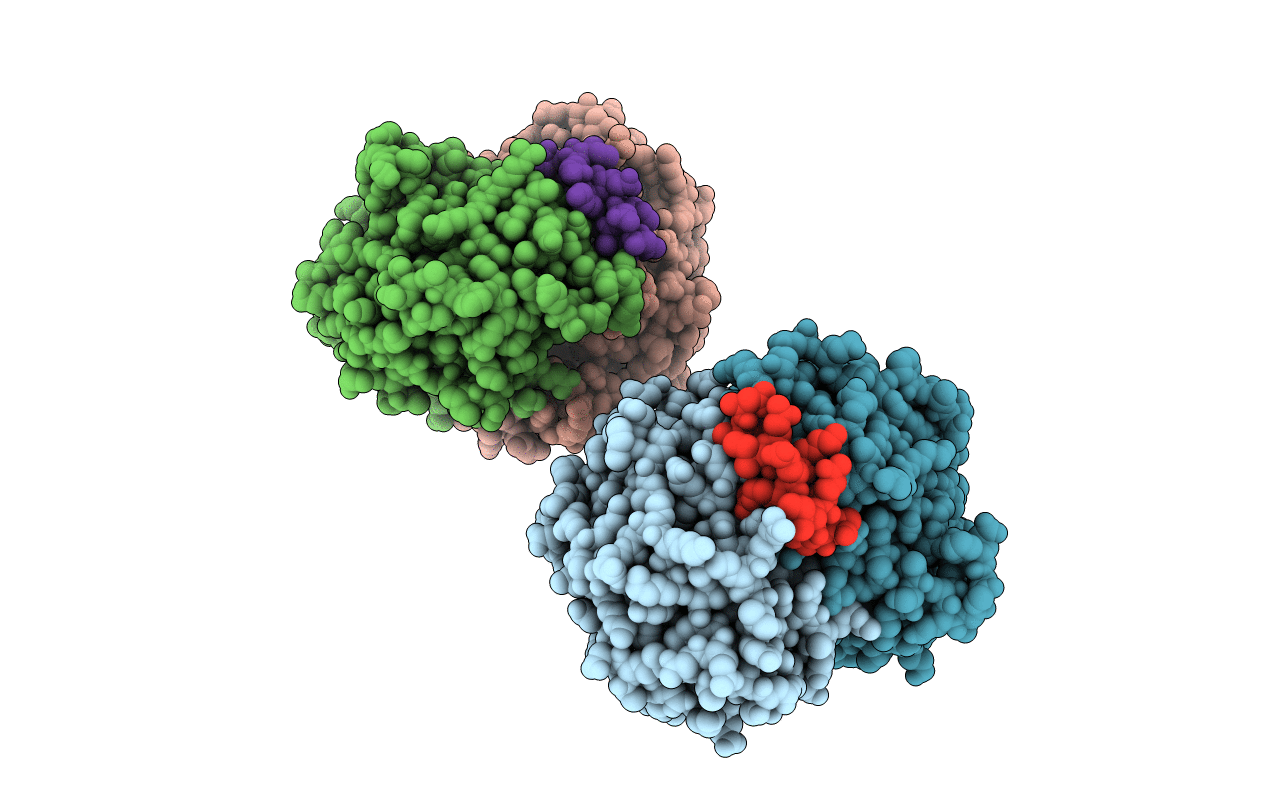 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-08-17 Classification: CELL CYCLE,HYDROLASE/CELL CYCLE Ligands: ADP |
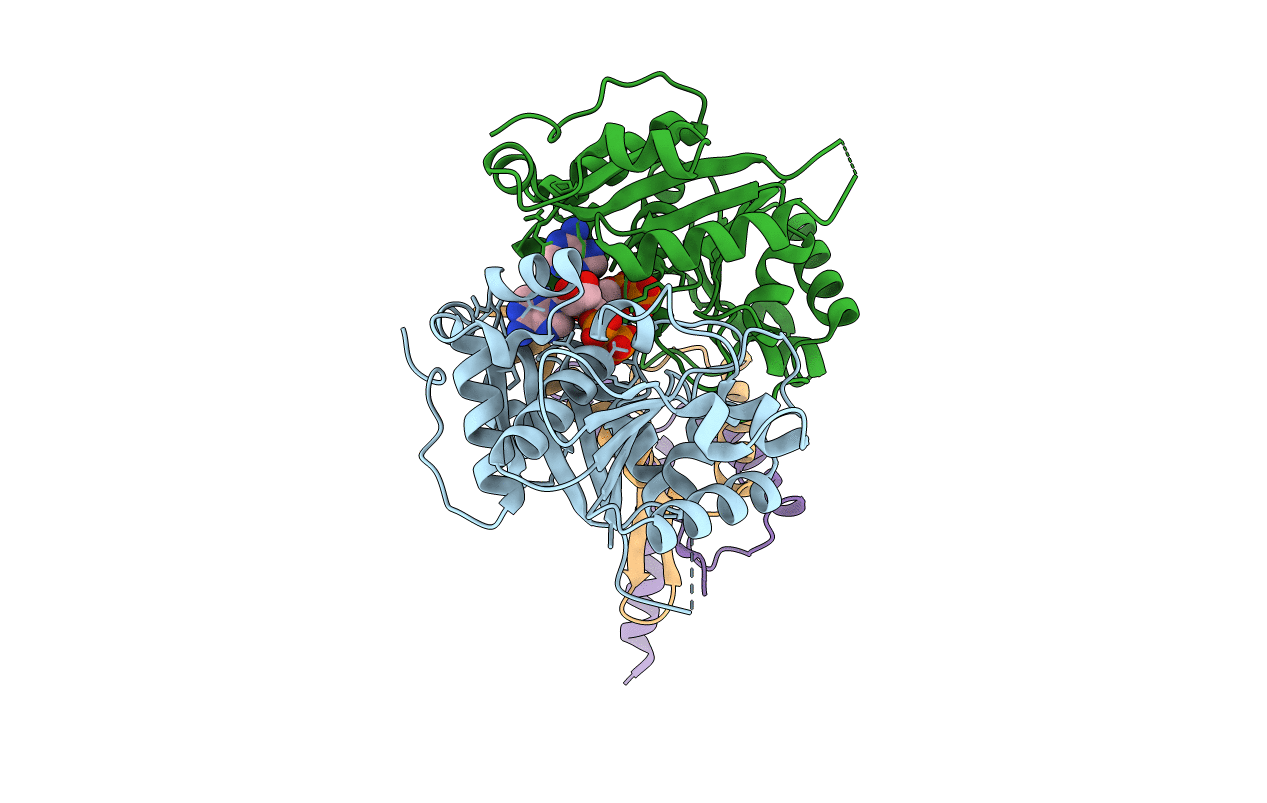 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:4.30 Å Release Date: 2011-08-17 Classification: CELL CYCLE,HYDROLASE/CELL CYCLE Ligands: ADP |
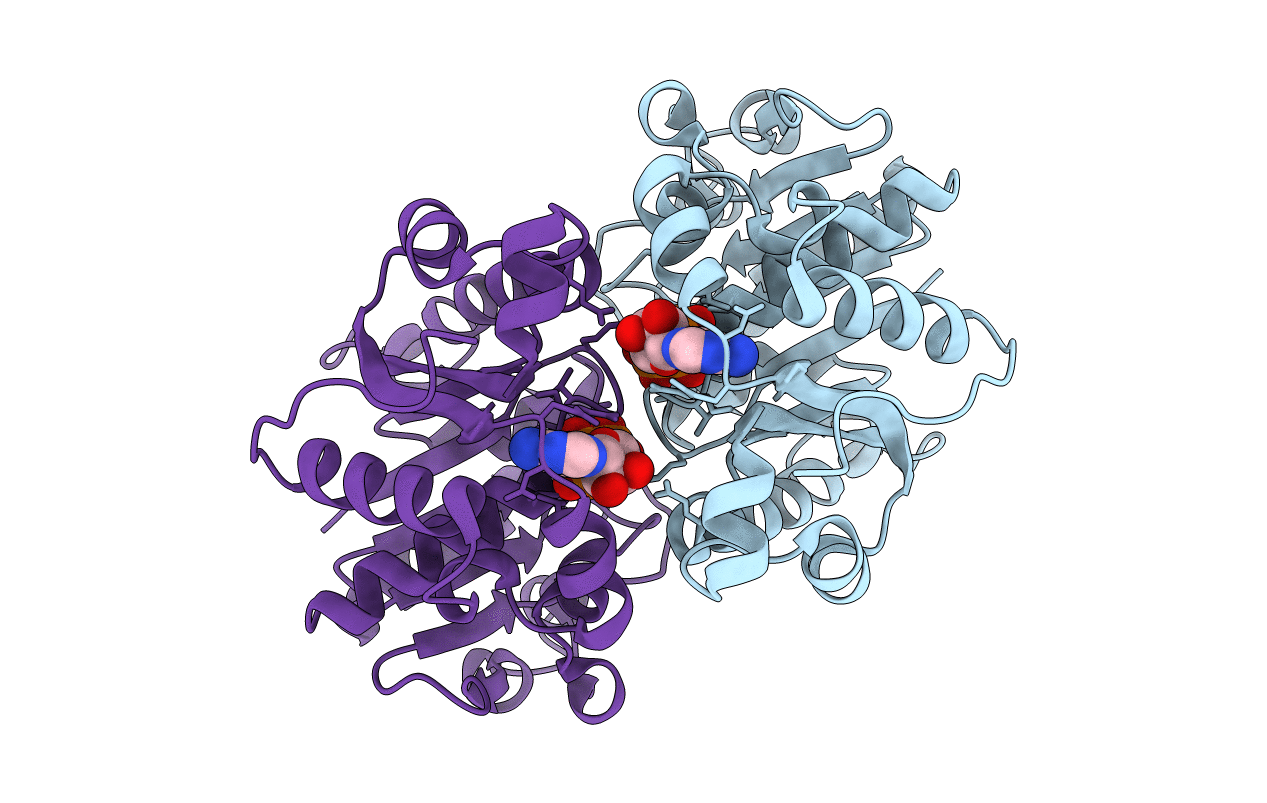 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2011-01-26 Classification: CELL CYCLE, HYDROLASE Ligands: MG, ATP |

