Search Count: 24
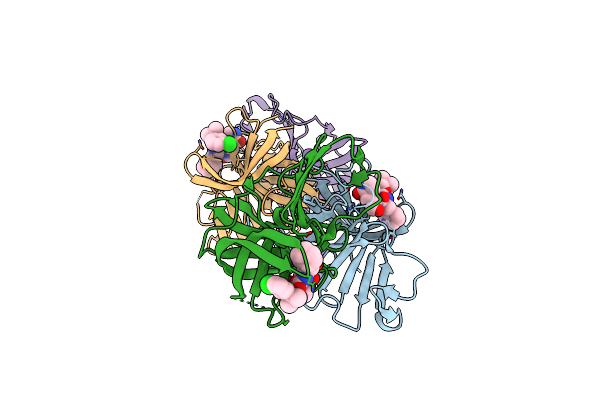 |
1.85 A Resolution Structure Of Norovirus 3Cl Protease In Complex With Inhibitor 5D
Organism: Norwalk virus (strain gi/human/united states/norwalk/1968)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-09-30 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: TKP |
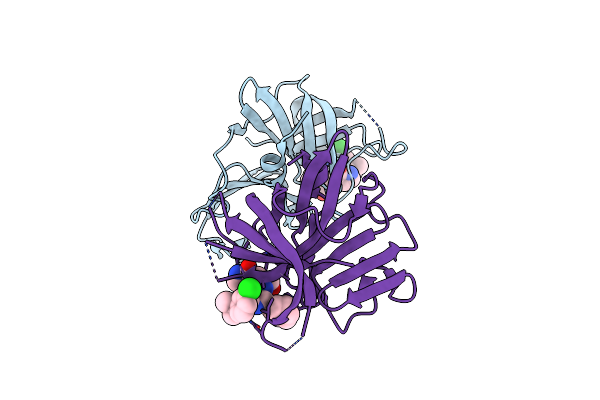 |
1.85 A Resolution Structure Of Norovirus 3Cl Protease In Complex With Inhibitor 7D
Organism: Norwalk virus (strain gi/human/united states/norwalk/1968)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-09-30 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: TKS |
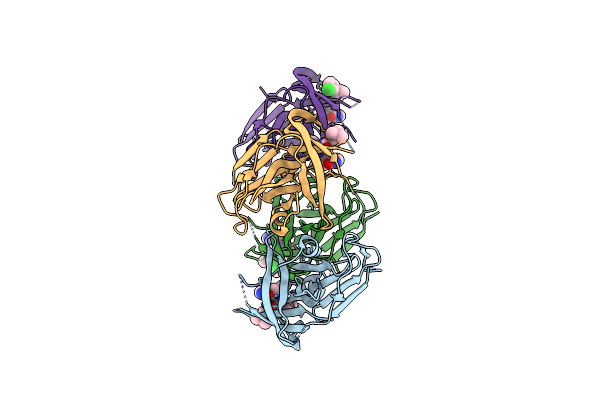 |
1.95 A Resolution Structure Of Norovirus 3Cl Protease In Complex With Inhibitor 5G
Organism: Norwalk virus (strain gi/human/united states/norwalk/1968)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-09-30 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: TKV |
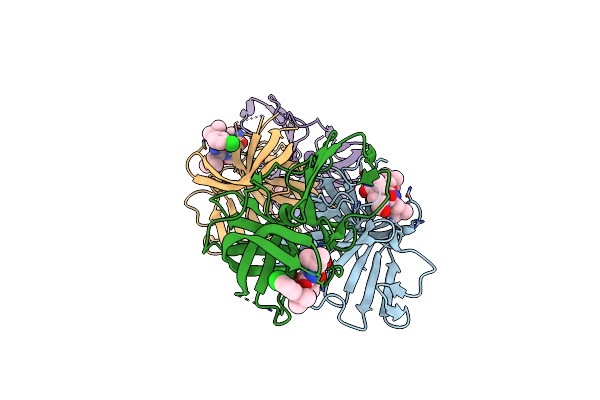 |
2.1 A Resolution Structure Of Norovirus 3Cl Protease In Complex With Inhibitor 7G
Organism: Norwalk virus (strain gi/human/united states/norwalk/1968)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-09-30 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: T0D |
 |
2.05 A Resolution Structure Of Mers 3Cl Protease In Complex With Inhibitor Gc376
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-04-04 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, B1S, K36 |
 |
1.55 A Resolution Structure Of Mers 3Cl Protease In Complex With Inhibitor Gc813
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-04-04 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: PG4, MG, AW4, B3G |
 |
1.85 A Resolution Structure Of Mers 3Cl Protease In Complex With Piperidine-Based Peptidomimetic Inhibitor 17
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-04-04 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: AVY, B3J |
 |
2.25 A Resolution Structure Of Mers 3Cl Protease In Complex With Piperidine-Based Peptidomimetic Inhibitor 21
Organism: Middle east respiratory syndrome-related coronavirus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-04-04 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: N02, B6Y |
 |
1.95 A Resolution Structure Of Norovirus 3Cl Protease In Complex With A Dipeptidyl Oxazolidinone-Based Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-12-13 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: V45 |
 |
1.40 A Resolution Structure Of Norovirus 3Cl Protease In Complex With The A M-Chlorophenyl Substituted Macrocyclic Inhibitor (17-Mer)
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-01-11 Classification: PROTEASE/PROTEASE INHIBITOR Ligands: CL, V56 |
 |
1.75 A Resolution Structure Of Norovirus 3Cl Protease In Complex With The A N-Pentyl Substituted Macrocyclic Inhibitor (17-Mer)
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-01-11 Classification: PROTEASE/PROTEASE INHIBITOR Ligands: V69 |
 |
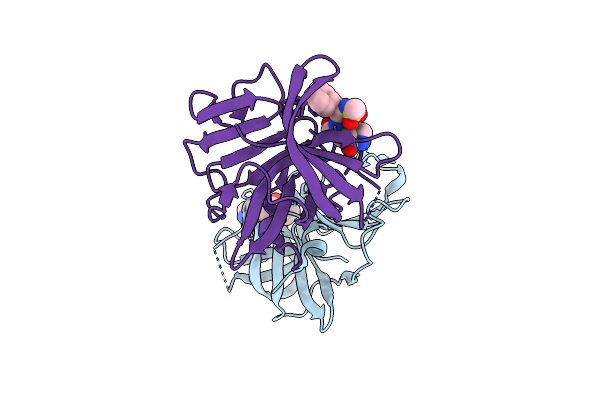
2.10 A Resolution Structure Of Norovirus 3Cl Protease In Complex With The Dipeptidyl Inhibitor 7L (Hexagonal Form)
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-11-23 Classification: PROTEASE/PROTEASE INHIBITOR Ligands: N38 |
 |
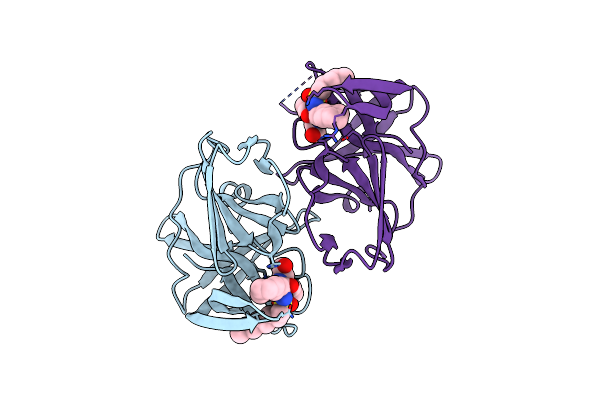
1.90 A Resolution Structure Of Norovirus 3Cl Protease In Complex With The Dipeptidyl Inhibitor 7L (Orthorhombic P Form)
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-23 Classification: PROTEASE/PROTEASE INHIBITOR Ligands: N38 |
 |
2.45 A Resolution Structure Of Norovirus 3Cl Protease In Complex With The Dipeptidyl Inhibitor 7M (Hexagonal Form)
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-11-23 Classification: PROTEASE/PROTEASE INHIBITOR Ligands: N40 |
 |
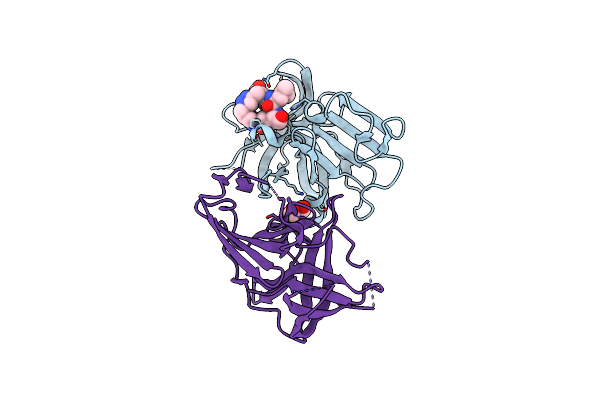
1.20 A Resolution Structure Of Norovirus 3Cl Protease In Complex With A Triazole-Based Macrocyclic (17-Mer) Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-05-04 Classification: PROTEASE/PROTEASE INHIBITOR Ligands: CL, 5LG |
 |
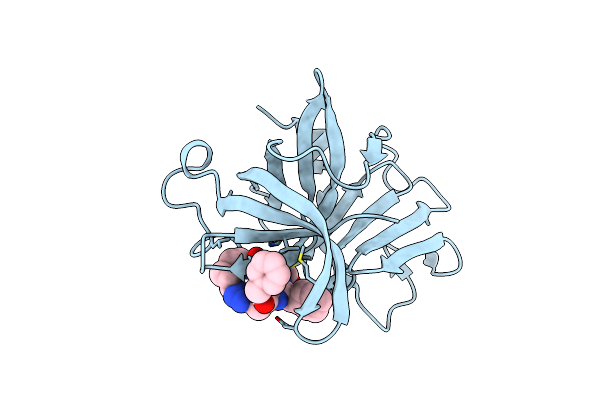
1.95 A Resolution Structure Of Norovirus 3Cl Protease In Complex With A Triazole-Based Macrocyclic (18-Mer) Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-05-04 Classification: PROTEASE/PROTEASE INHIBITOR Ligands: 5LH, GOL |
 |
1.20 A Resolution Structure Of Norovirus 3Cl Protease In Complex With A Triazole-Based Macrocyclic (21-Mer) Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-05-04 Classification: PROTEASE/PROTEASE INHIBITOR Ligands: CL, 5LJ |
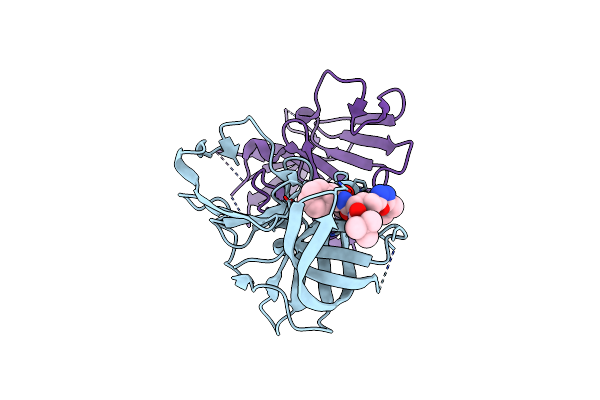 |
2.35A Resolution Structure Of Norovirus 3Cl Protease In Complex An Oxadiazole-Based, Cell Permeable Macrocyclic (21-Mer) Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-02-10 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: CL, V66 |
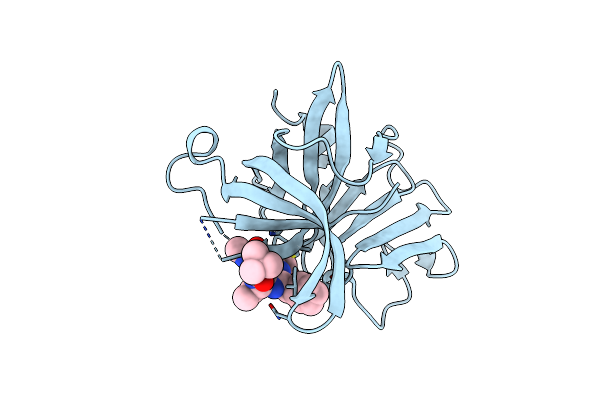 |
1.0A Resolution Structure Of Norovirus 3Cl Protease In Complex An Oxadiazole-Based, Cell Permeable Macrocyclic (20-Mer) Inhibitor
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2016-02-10 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: V64 |
 |
Organism: Pimelobacter simplex
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-05-26 Classification: OXIDOREDUCTASE Ligands: SF4, FMN, ADP |

