Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
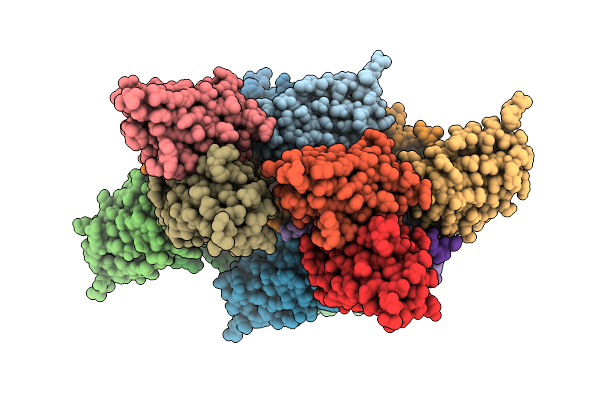 |
Cryo-Em Structure Of Z-Dna Binding Antibody Z-D11 In Complex With Left-Handed Z-Dna
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN Ligands: MG |
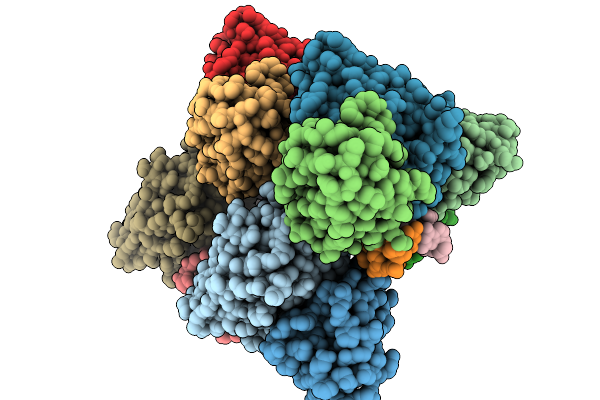 |
Cryo-Em Structure Of Z22 Mab In Complex With Left-Handed Z-Dna (Dimer Of Trimer)
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN Ligands: MG |
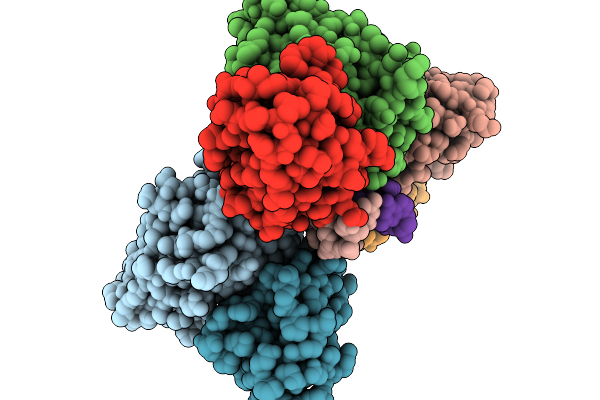 |
Cryo-Em Structure Of Z22 Antibody In Complex With Left-Handed Z-Dna (Trimer)
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
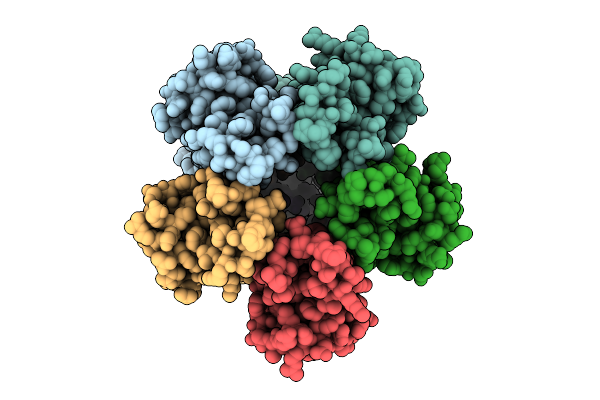 |
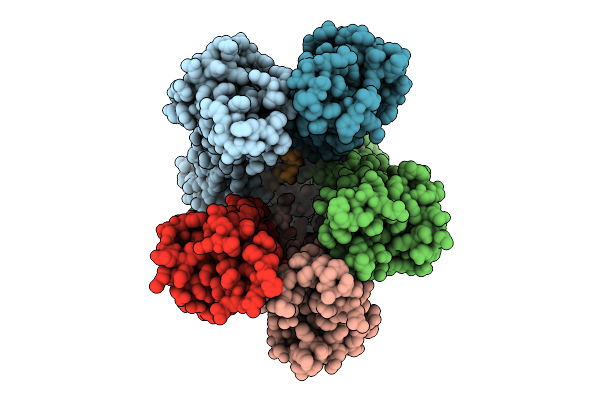
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
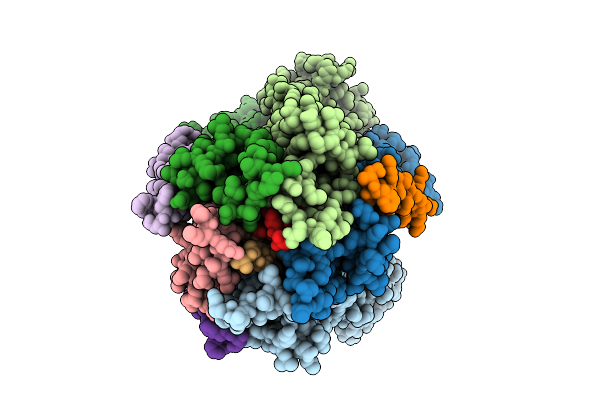
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
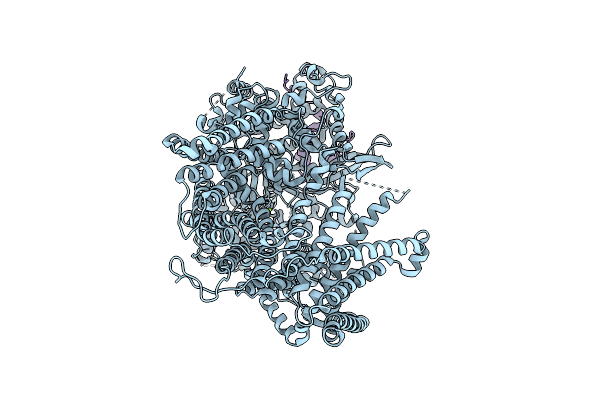
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
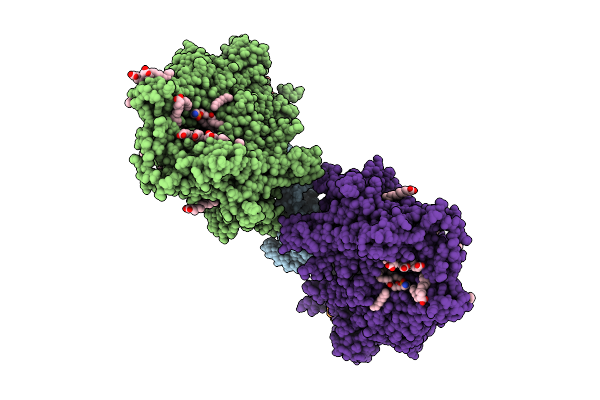
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG, DD9, D10, HP6, XKP, PEF, PLM, DCR |
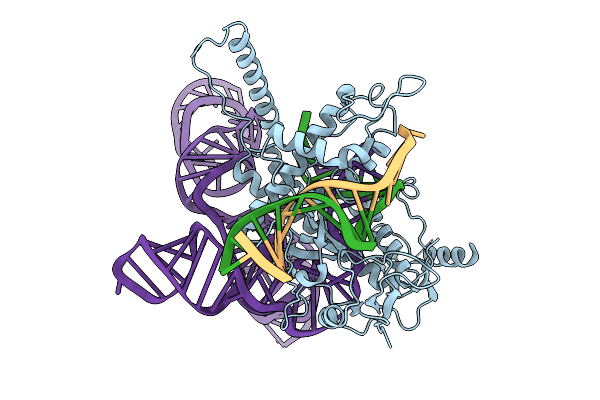 |
Structure Of Latranc Complex Bound To 27Nt Complementary Dna Substrate, Conformation 1
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
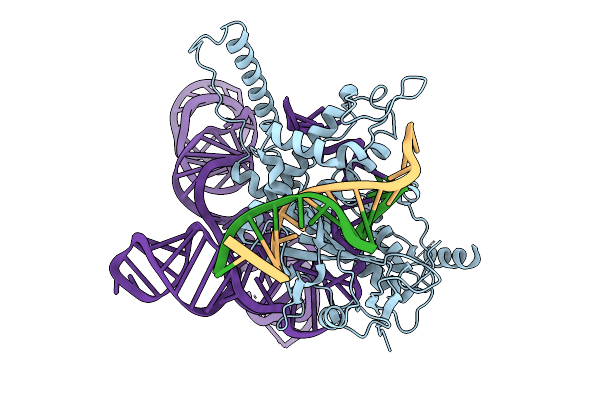 |
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
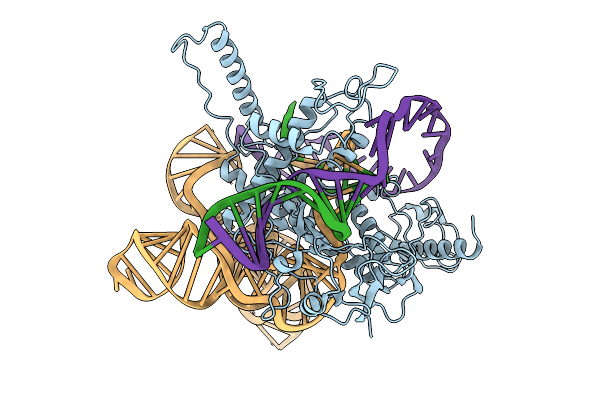 |
Structure Of Latranc Complex Bound To 27Nt Complementary Dna Substrate, Conformation 2
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
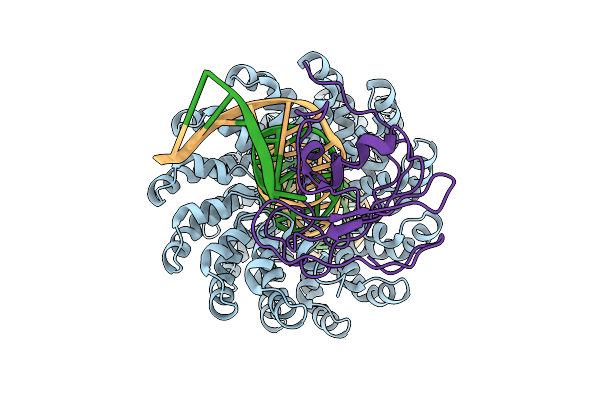 |
Organism: Xanthomonas, Burkholderia cenocepacia h111, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
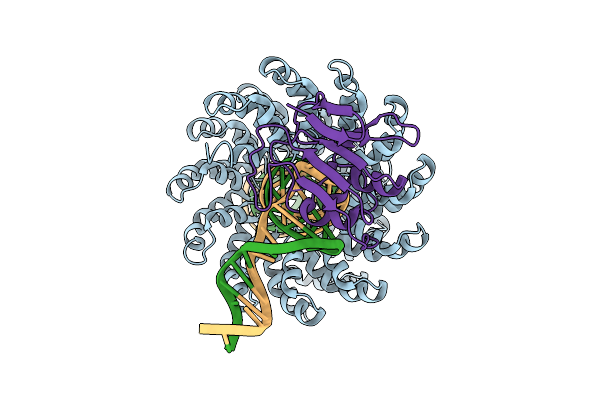 |
Organism: Xanthomonas, Burkholderia cenocepacia h111, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
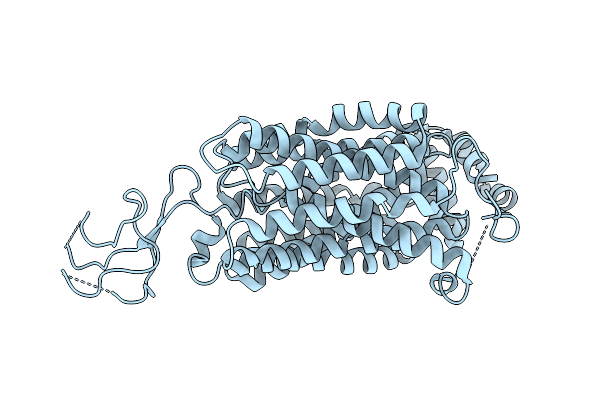 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN |
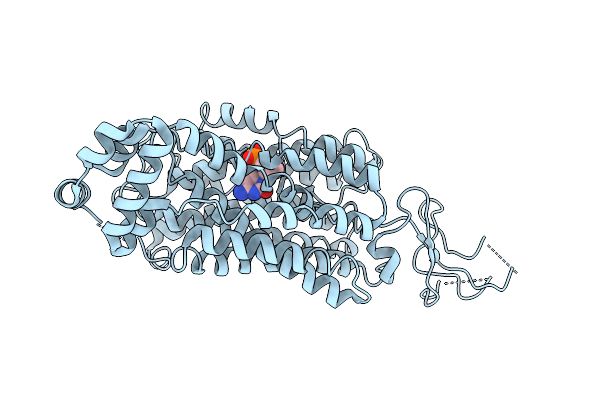 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: L8P |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: GBM |
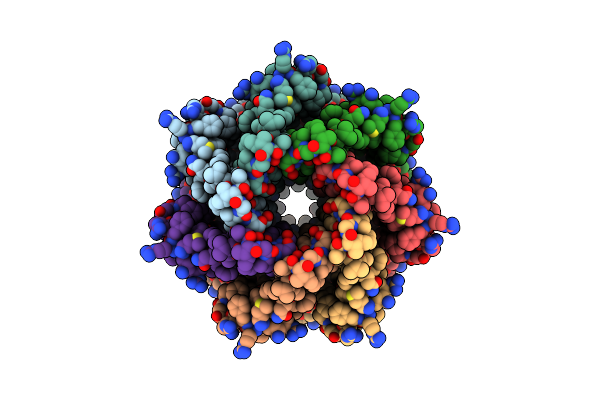 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: TRANSPORT PROTEIN |
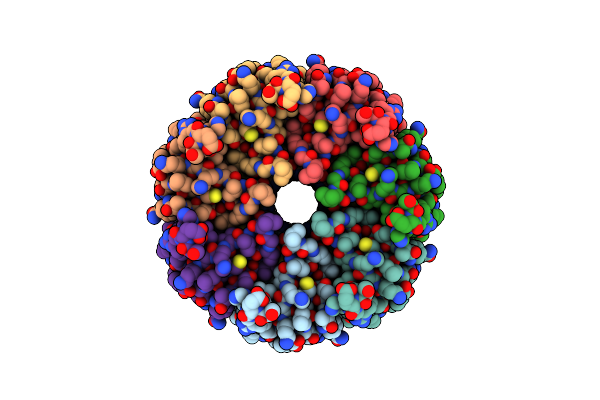 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: TRANSPORT PROTEIN |
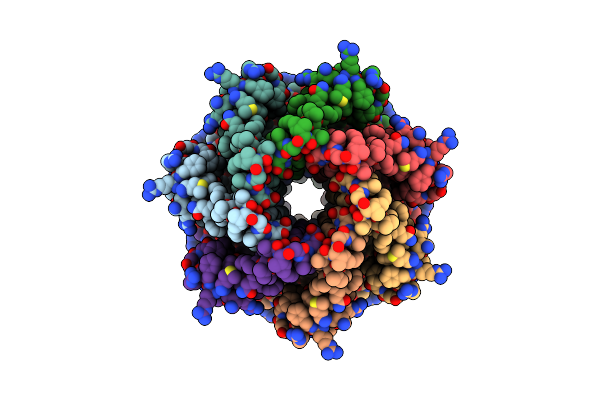 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: TRANSPORT PROTEIN |
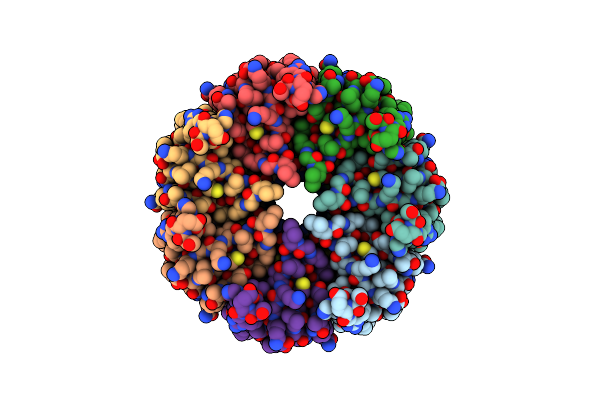 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: TRANSPORT PROTEIN |