Search Count: 317
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG |
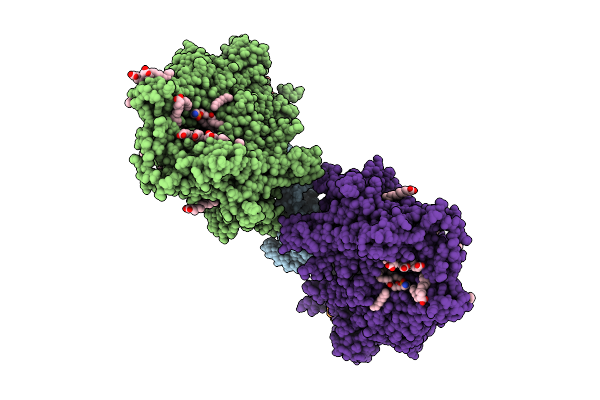 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG, DD9, D10, HP6, XKP, PEF, PLM, DCR |
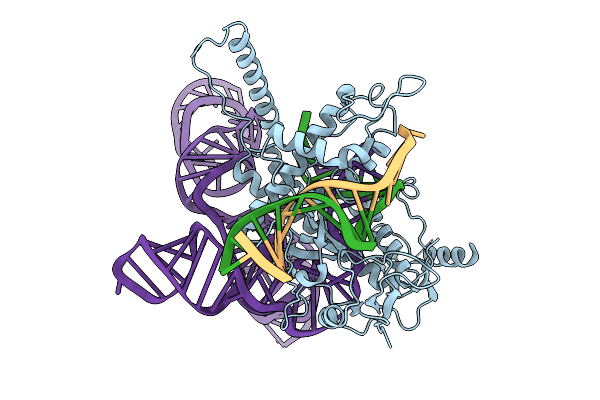 |
Structure Of Latranc Complex Bound To 27Nt Complementary Dna Substrate, Conformation 1
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
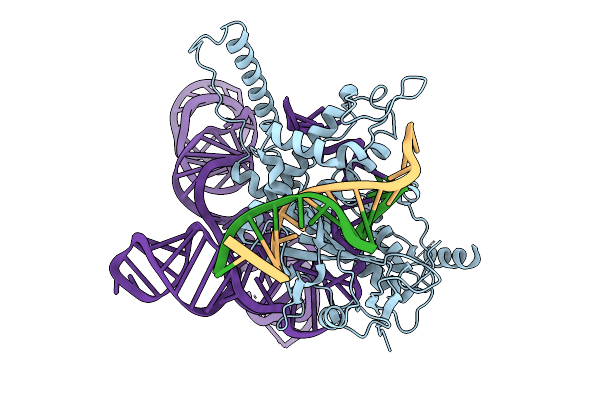 |
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
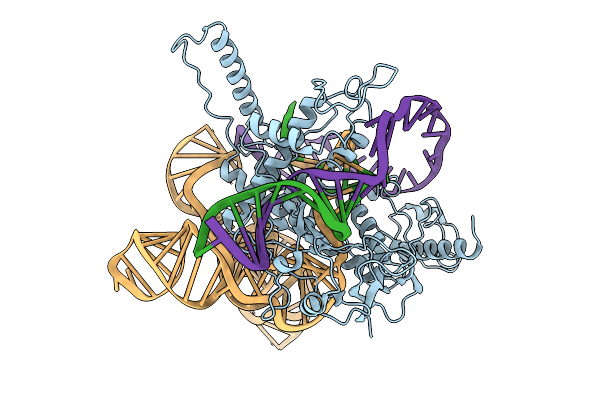 |
Structure Of Latranc Complex Bound To 27Nt Complementary Dna Substrate, Conformation 2
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
 |
Organism: Xanthomonas, Burkholderia cenocepacia h111, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Xanthomonas, Burkholderia cenocepacia h111, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: L8P |
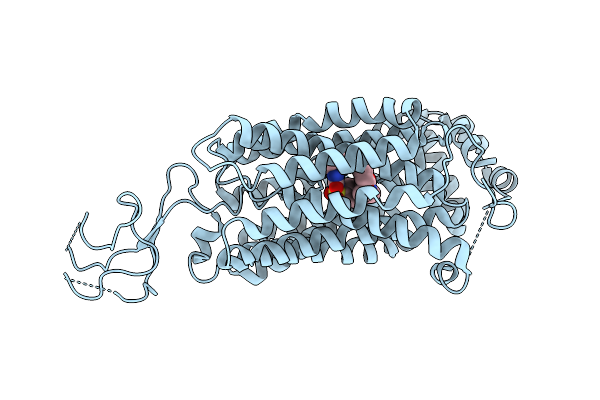 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: GBM |
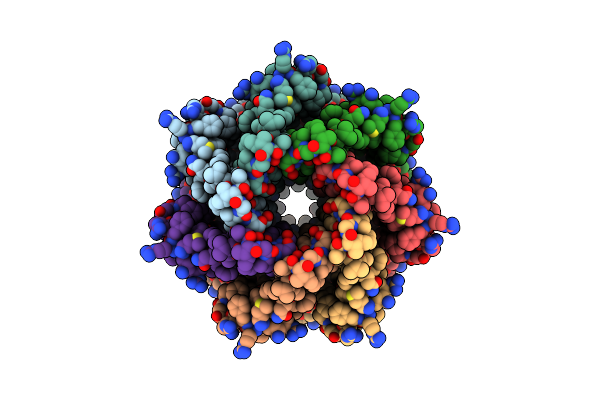 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: TRANSPORT PROTEIN |
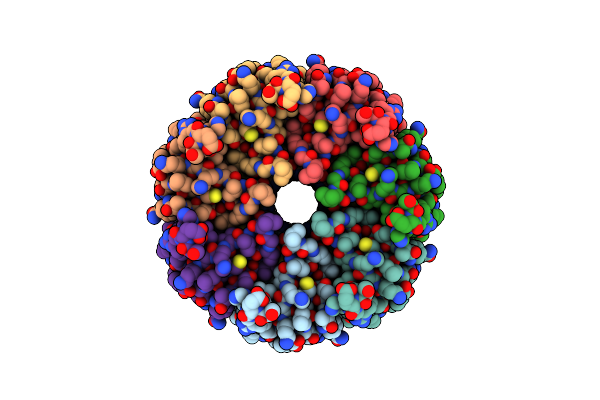 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: TRANSPORT PROTEIN |
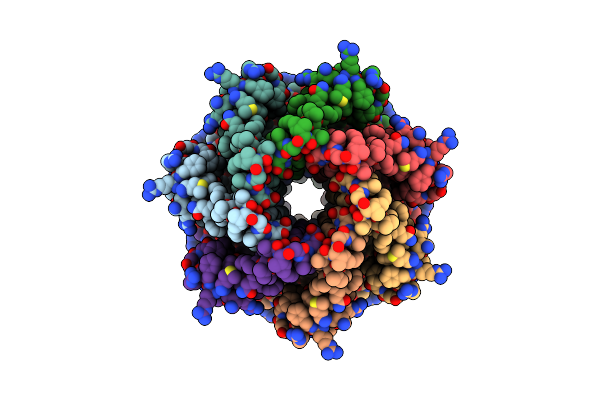 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: DNA Ligands: TEP, K, IOD, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: DNA Ligands: HPA, NA, K |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: DNA Ligands: I7U, NA |

