Search Count: 74
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: TRANSPORT PROTEIN Ligands: Z41, PLM |
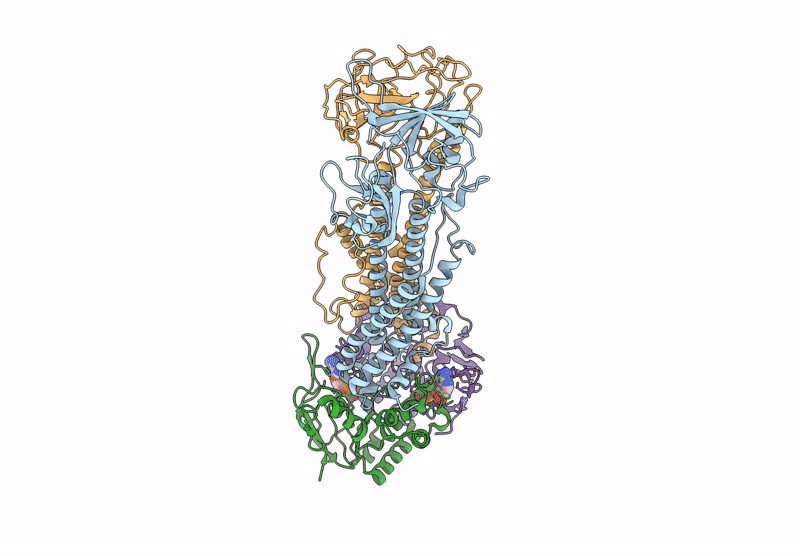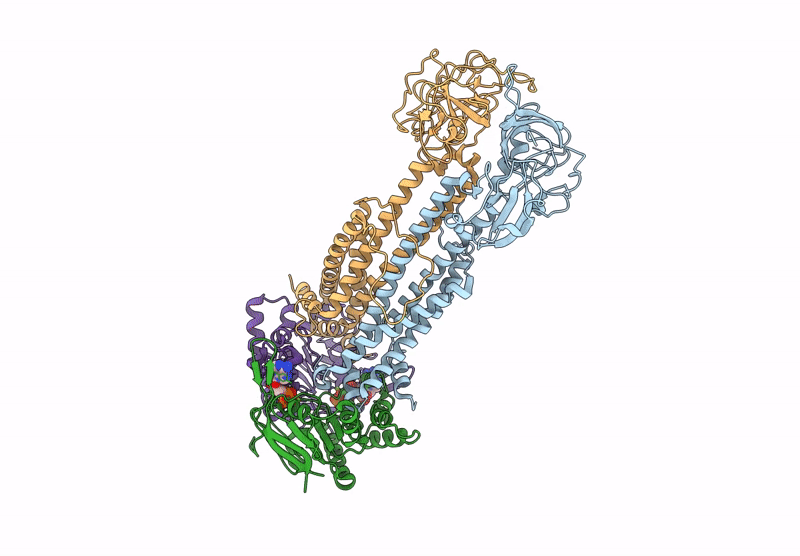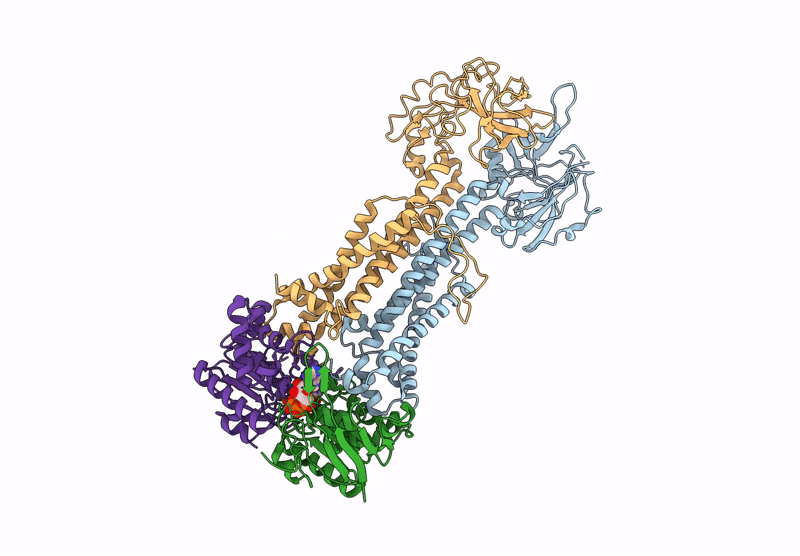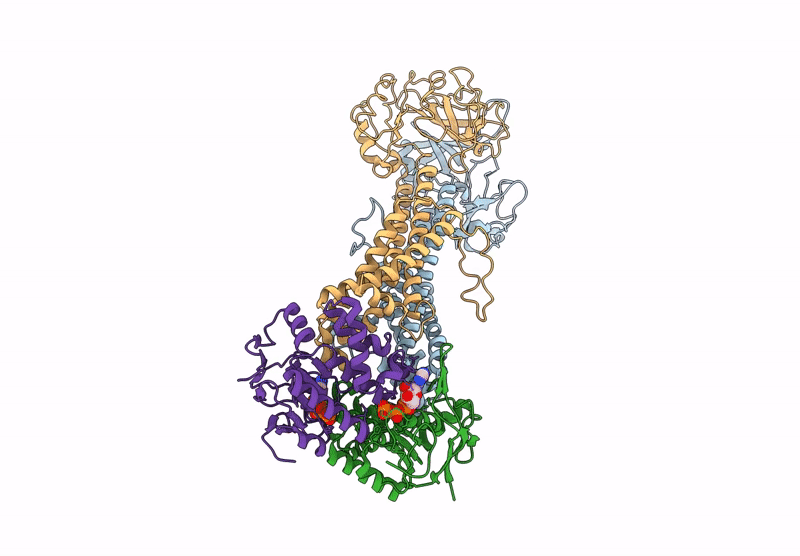 |
Cryo-Em Structure Of Endogenous Atp-Bound Lolcde With Lold-E171Q Mutations In Nanodiscs
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: TRANSPORT PROTEIN Ligands: MG, ATP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-09-18 Classification: HYDROLASE Ligands: GOL, SO4, NA |
 |
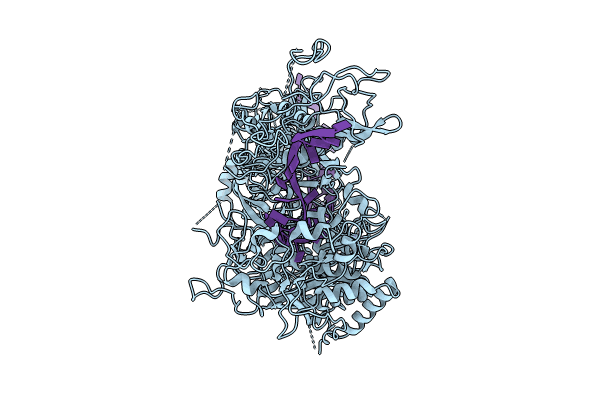
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-09-18 Classification: HYDROLASE Ligands: NAD, SO4 |
 |
Organism: Desulfonema ishimotonii, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Organism: Desulfonema ishimotonii, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
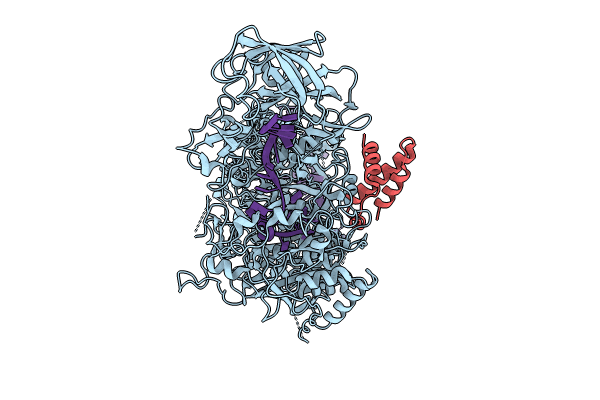
Organism: Escherichia coli, Desulfonema ishimotonii
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
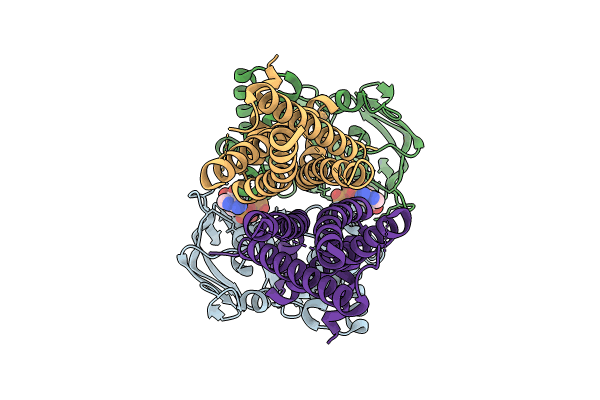
Organism: Escherichia coli, Desulfonema ishimotonii
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
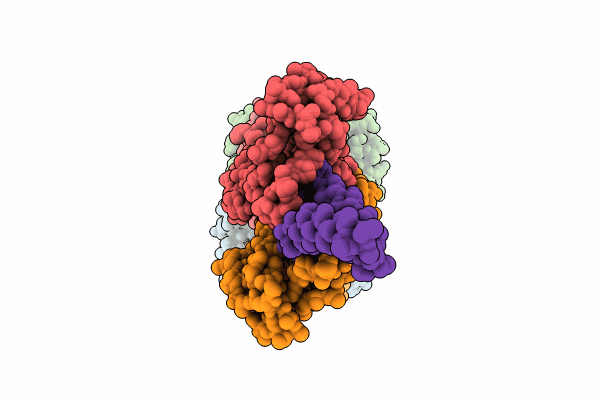
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSPORT PROTEIN Ligands: ATP |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: MEMBRANE PROTEIN Ligands: ATP |
 |
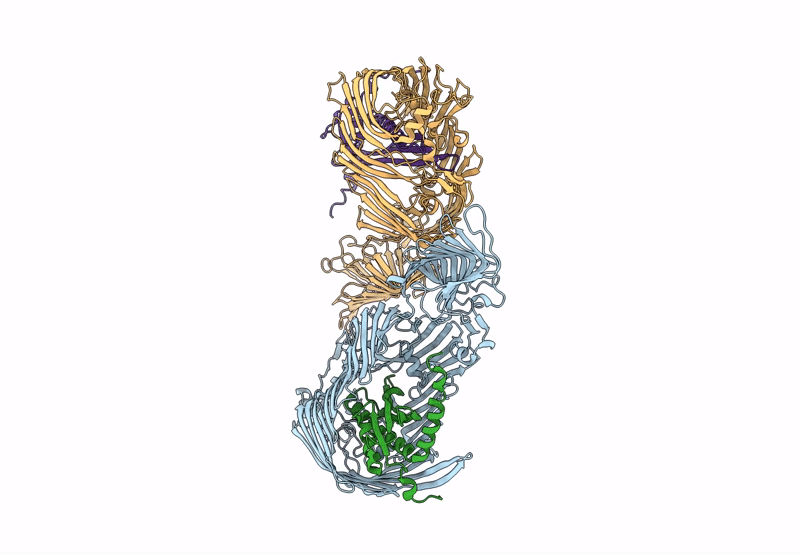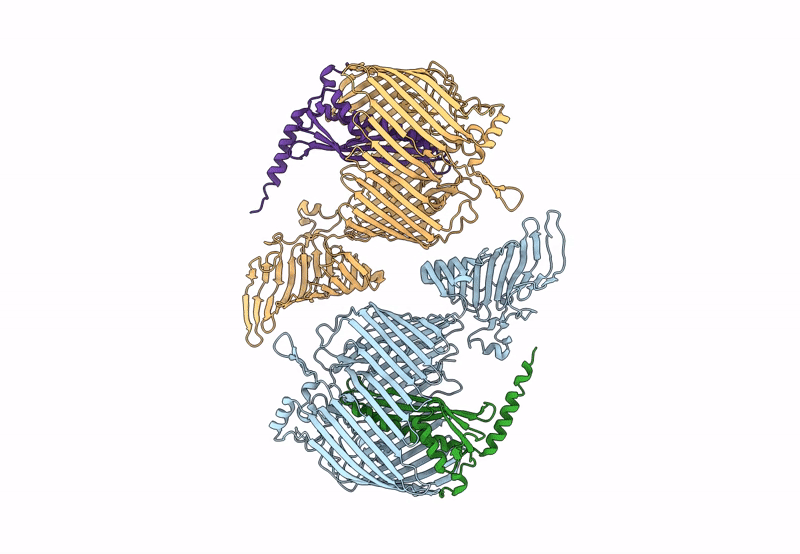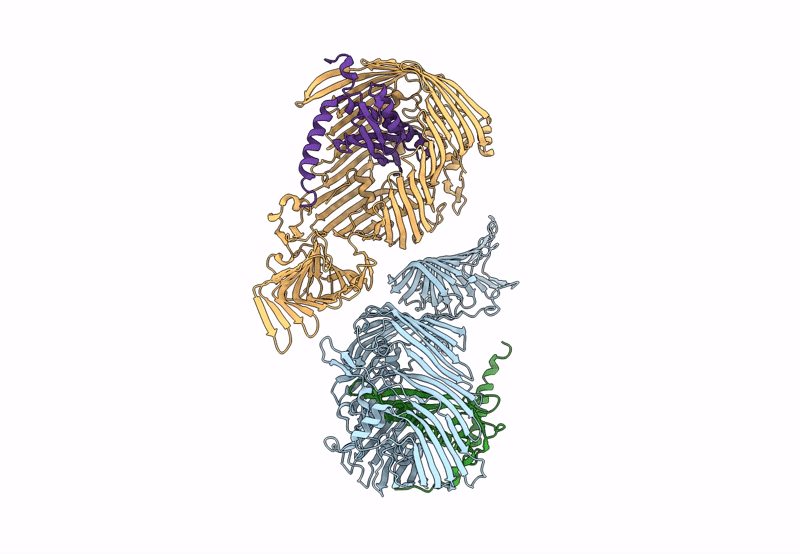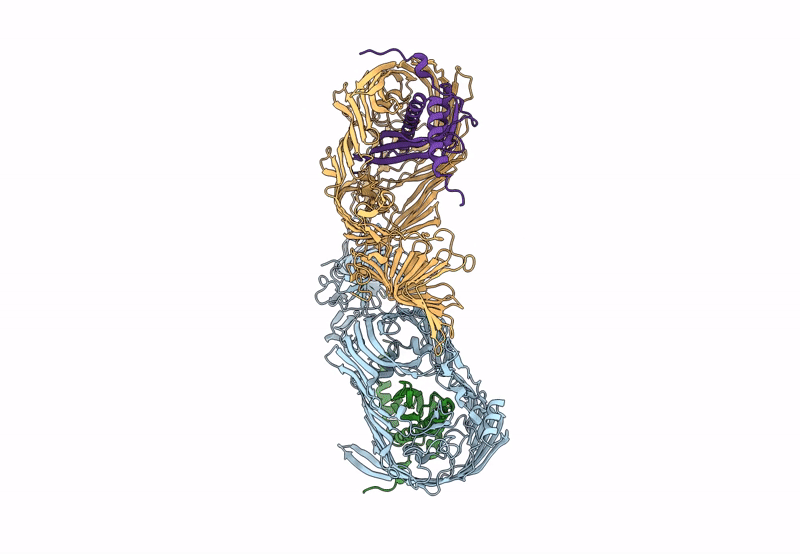
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: PCJ |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN |
 |
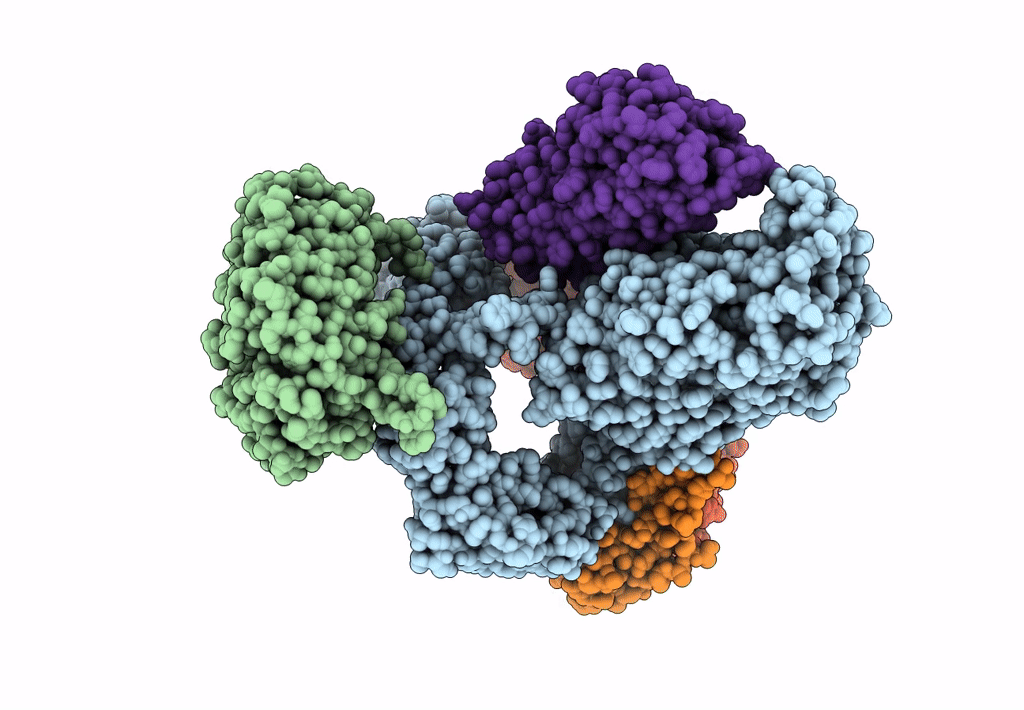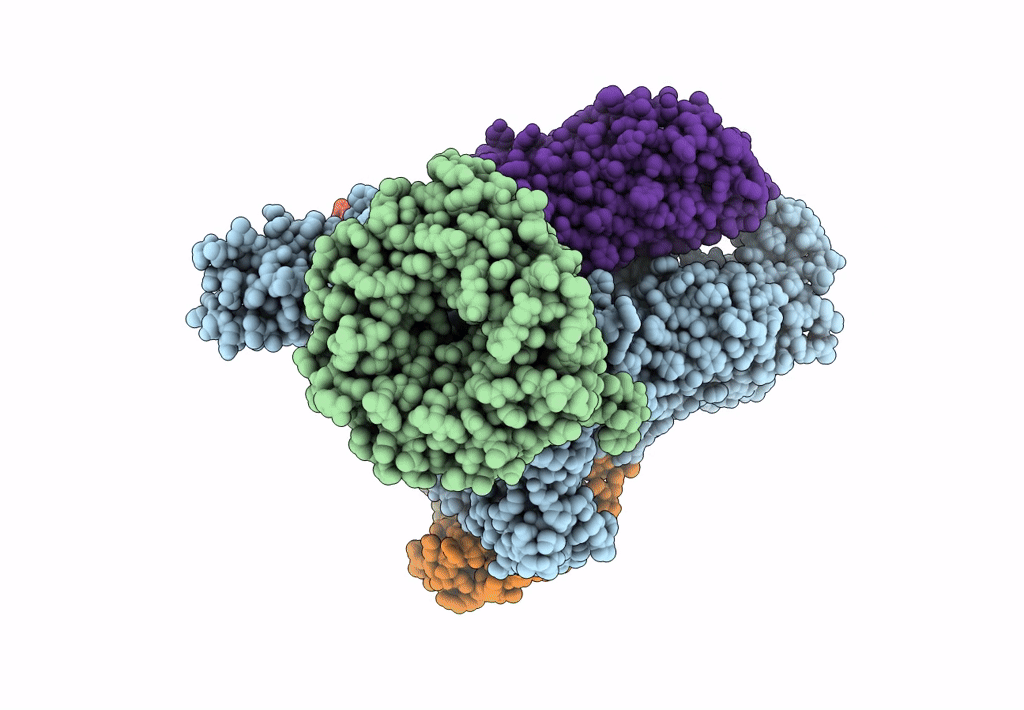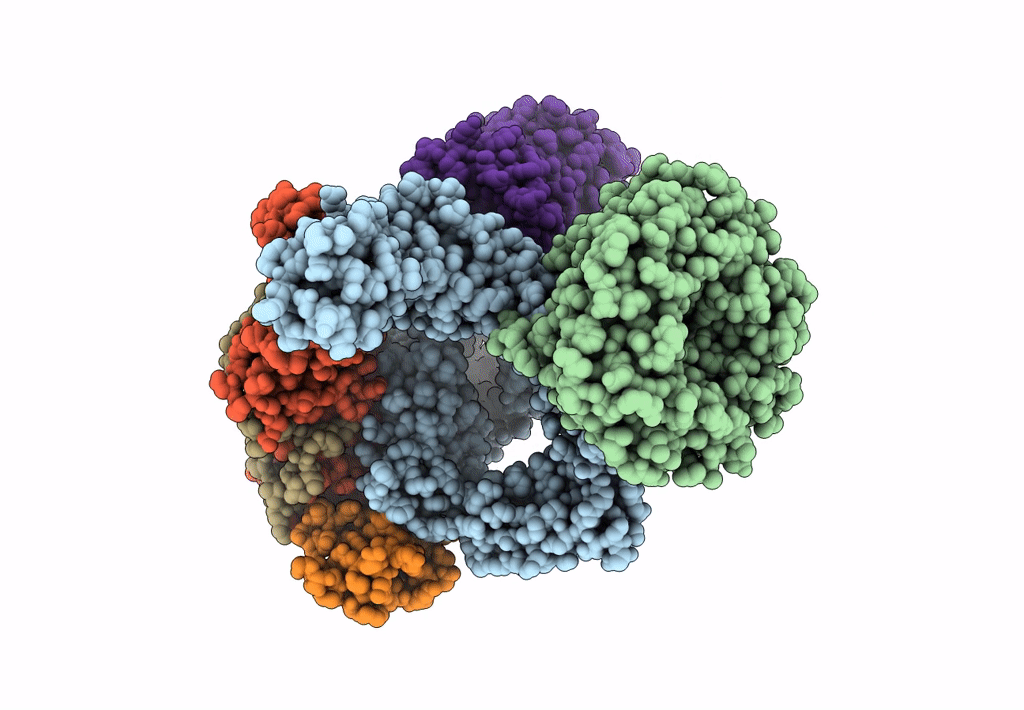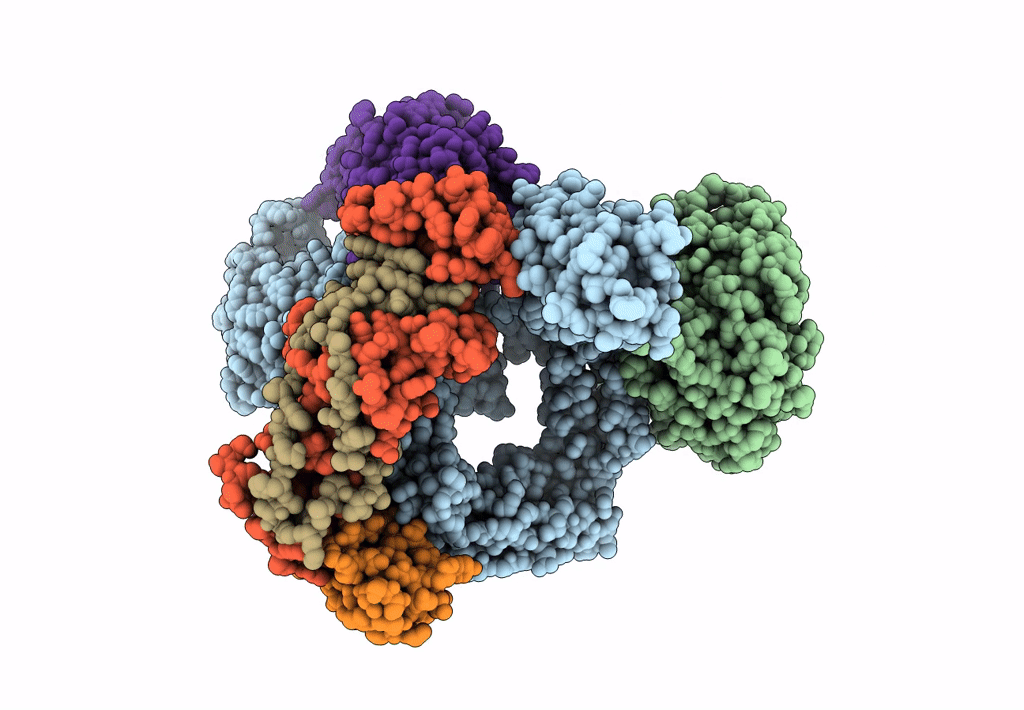
Bam-Espp Complex Structure With Bama-G431C And G781C/Espp-N1293C And A1043C Mutations In Nanodisc
Organism: Escherichia coli k-12, Escherichia coli o157:h7
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: MEMBRANE PROTEIN |
 |
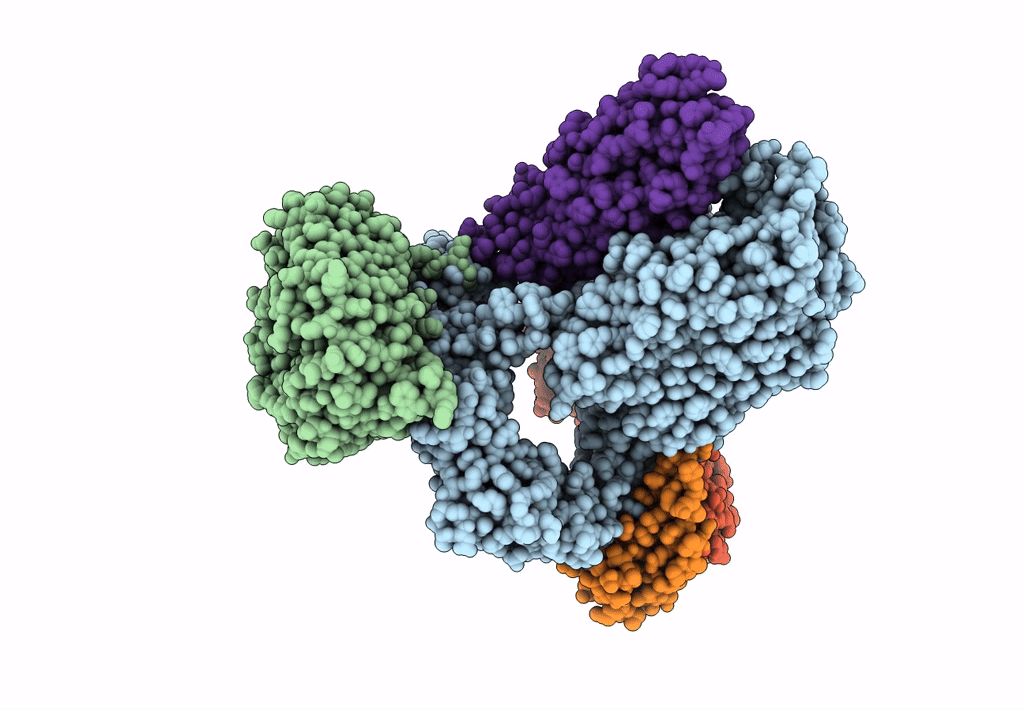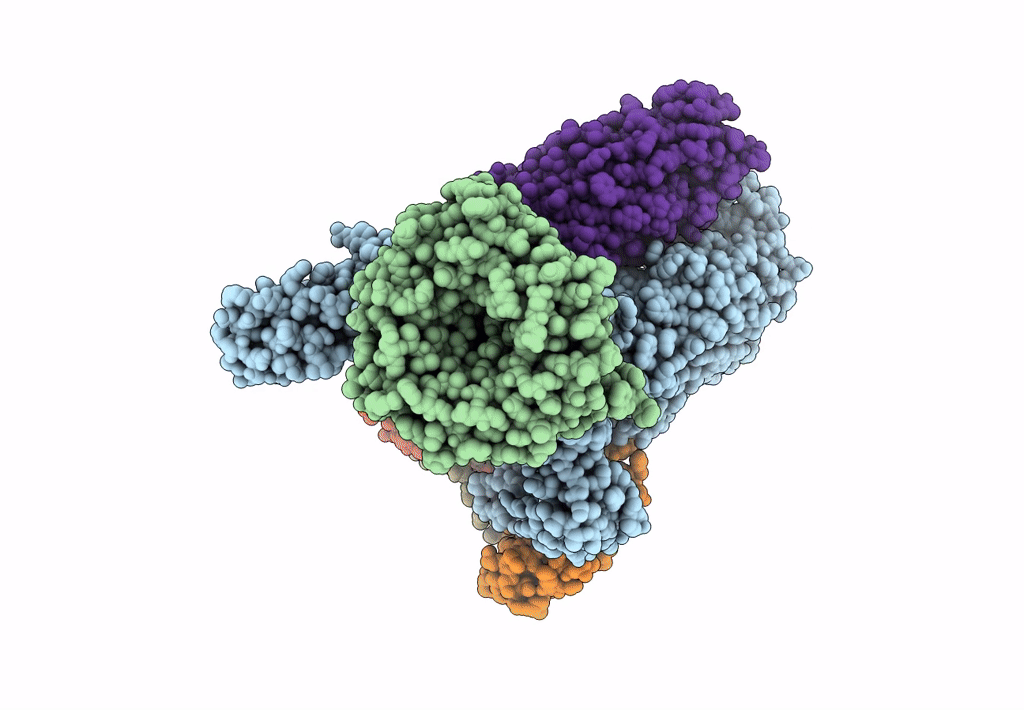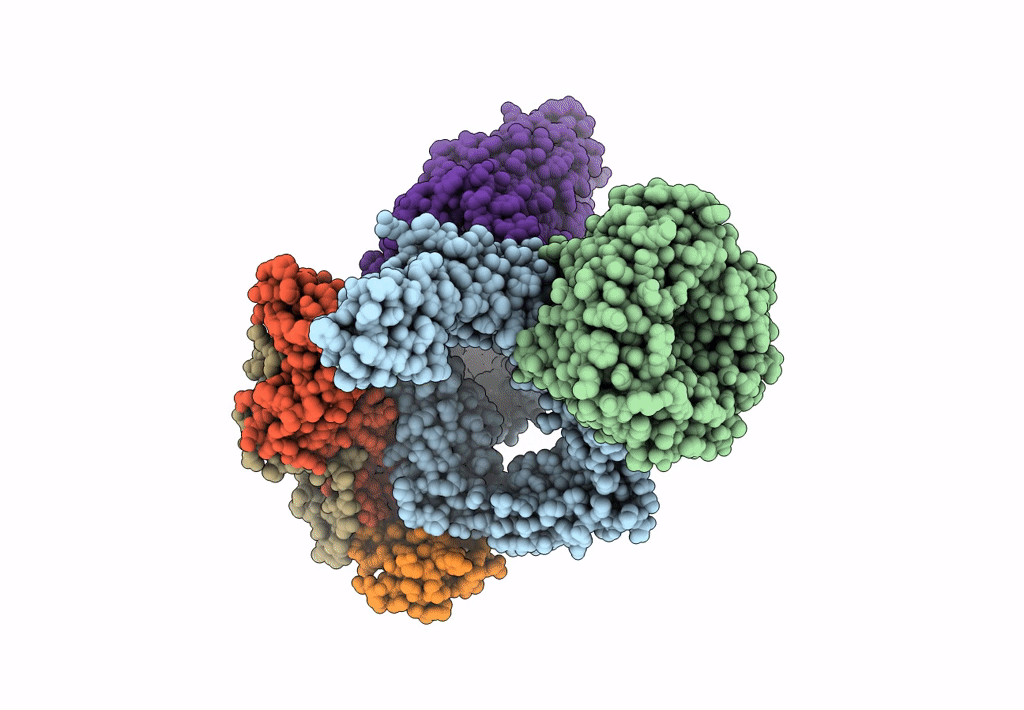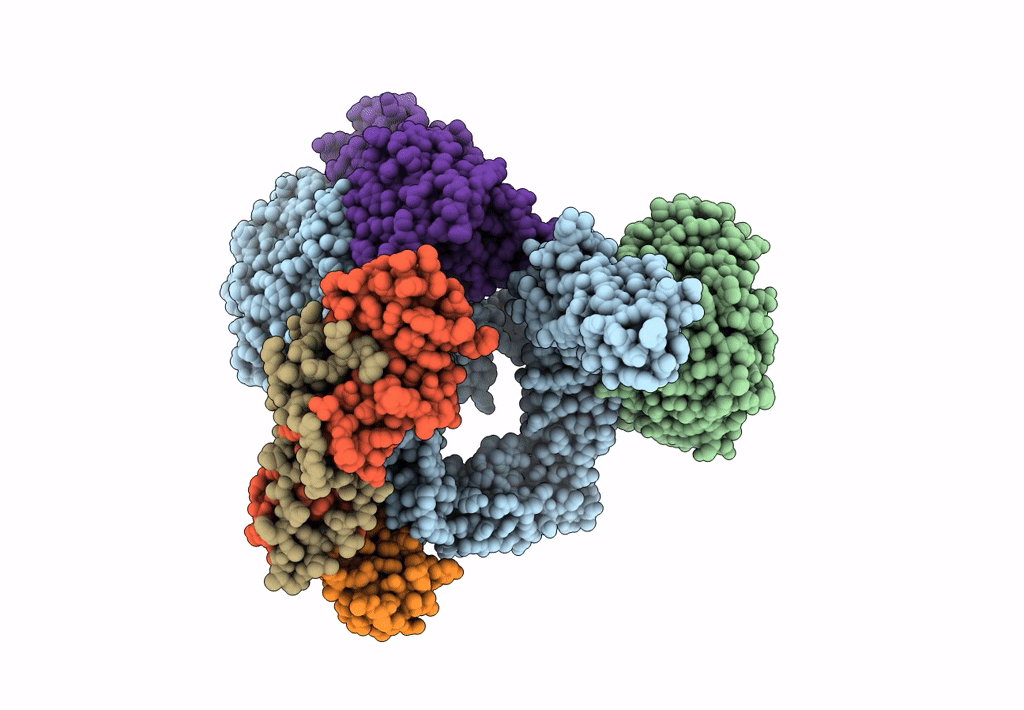
Bam-Espp Complex Structure With Bama-N427C/Espp-R1297C Mutations In Nanodisc
Organism: Escherichia coli k-12, Escherichia coli o157:h7
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: MEMBRANE PROTEIN |
 |
Bam-Espp Complex Structure With Bama-G431C/Espp-N1293C Mutations In Nanodisc
Organism: Escherichia coli k-12, Escherichia coli o157:h7
Method: ELECTRON MICROSCOPY Release Date: 2023-04-26 Classification: TRANSPORT PROTEIN |
 |
Bam-Espp Complex Structure With Bama-S425C/Espp-S1299C Mutations In Nanodisc
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-26 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: MEMBRANE PROTEIN Ligands: PCJ |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: MG, ANP |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-07-20 Classification: TRANSCRIPTION |

