Search Count: 248
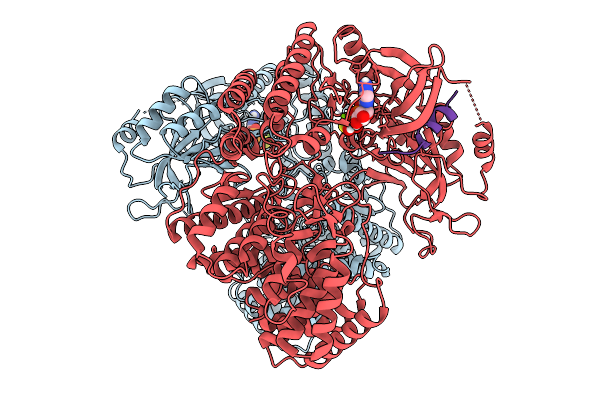 |
Cryo-Em Structure Of Classiii Salivaricin Modification Enzyme Salkc In The Presence Of Sala
Organism: Streptococcus salivarius
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: AGS, MG |
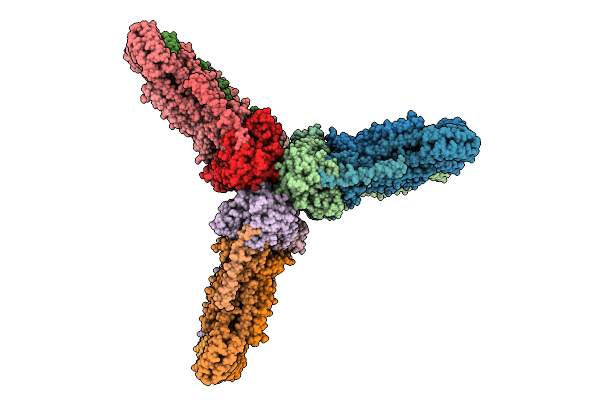 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: CELL CYCLE Ligands: AGS |
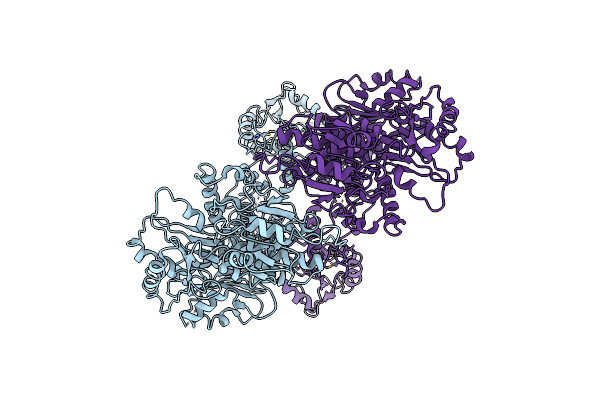 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
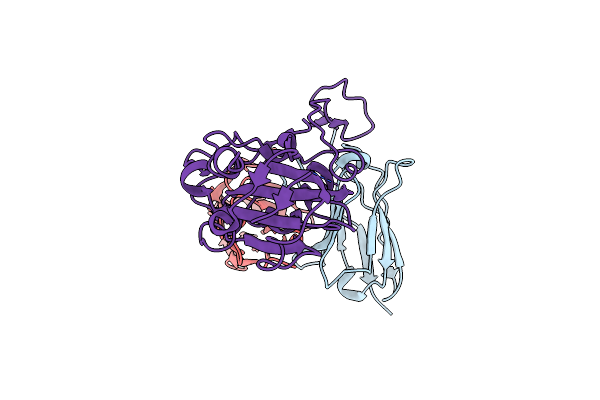 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
Organism: Bufavirus-1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRUS LIKE PARTICLE Ligands: SIA |
 |
Cryo-Em Structure Of Classiii Lanthipeptide Modification Enzyme Pnekc Mutant H522A.
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: ANTIMICROBIAL PROTEIN Ligands: PO4 |
 |
Cryo-Em Structure Of Classiii Lanthipeptide Modification Enzyme Pnekc In The Presence Of Gtp.
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: ANTIMICROBIAL PROTEIN Ligands: GTP |
 |
Cryo-Em Structure Of Classiii Lanthipeptide Modification Enzyme Pnekc With Chain A Bounded To Substrate Pnea And Gtp.
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: ANTIMICROBIAL PROTEIN Ligands: GTP, PO4 |
 |
Cryo-Em Structure Of Classiii Lanthipeptide Modification Enzyme Pnekc In The Presence Of Pnea And Gtprs.
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: ANTIMICROBIAL PROTEIN Ligands: MG, GSP, PO4 |
 |
Organism: Bufavirus-1
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Bufavirus-1
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Bufavirus-1
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2024-03-27 Classification: PROTEIN BINDING Ligands: VFI |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: MEMBRANE PROTEIN Ligands: ADP, MG, PO4 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: MEMBRANE PROTEIN Ligands: ADP, MG |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: MEMBRANE PROTEIN |

