Search Count: 14
 |
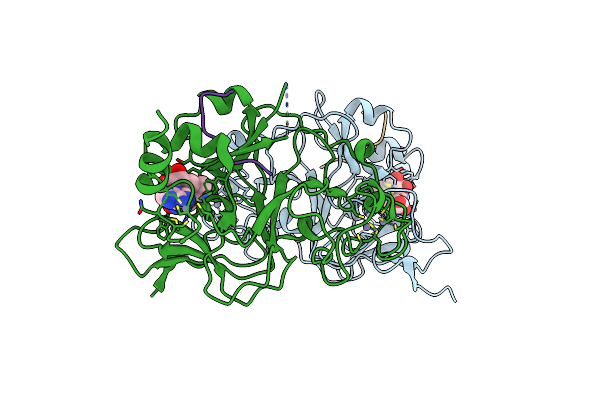
Structure Of G9A Set-Domain With Histone H3K9Norleucine Mutant Peptide And Bound S-Adenosylmethionine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2016-09-14 Classification: TRANSFERASE Ligands: ZN, SAM |
 |
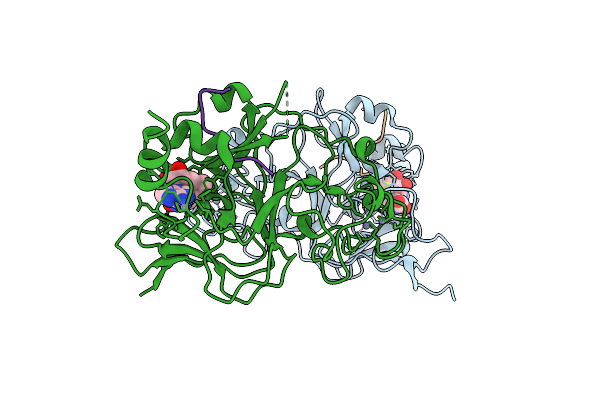
Structure Of G9A Set-Domain With Histone H3K9Ala Mutant Peptide And Bound S-Adenosylmethionine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2016-07-06 Classification: TRANSFERASE Ligands: ZN, SAM |
 |
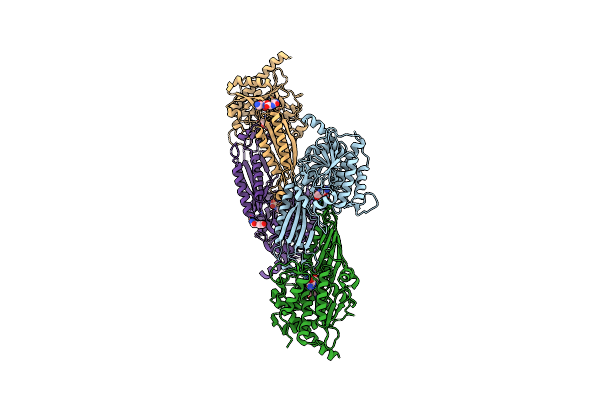
Structure Of G9A Set-Domain With Histone H3K9M Mutant Peptide And Bound S-Adenosylmethionine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-07-06 Classification: TRANSFERASE Ligands: ZN, SAM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2016-07-06 Classification: TRANSFERASE Ligands: ZN, SAM |
 |
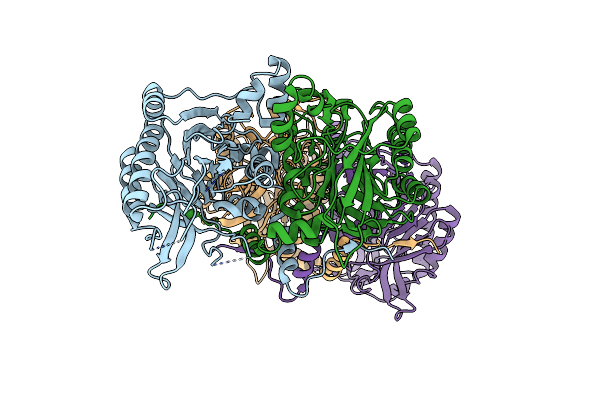
Crystal Structure Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With A Gly-Gly Peptide
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2008-05-13 Classification: HYDROLASE Ligands: GLY, ZN |
 |
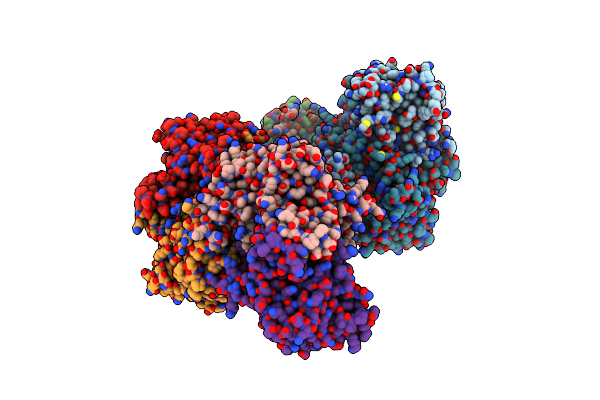
Crystal Structure Of A Pyrimidine Degrading Enzyme From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-03-25 Classification: HYDROLASE |
 |
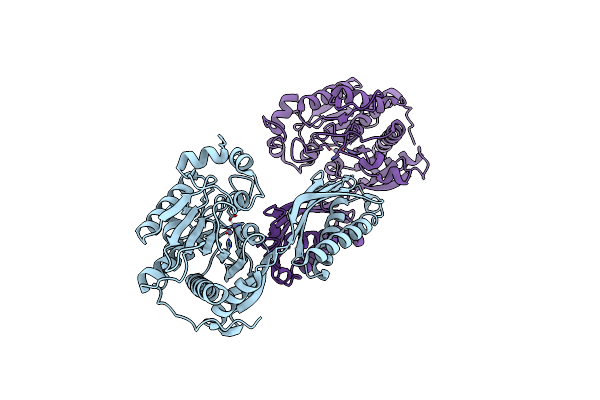
Crystal Structure Of A Pyrimidine Degrading Enzyme From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-03-25 Classification: HYDROLASE |
 |
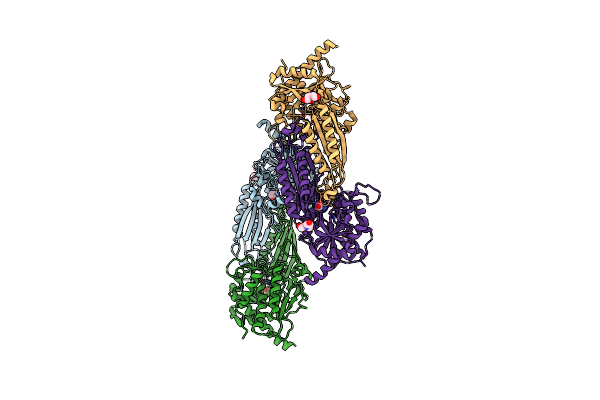
Crystal Structure Of Mutant E159A Of Beta-Alanine Synthase From Saccharomyces Kluyveri
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN |
 |
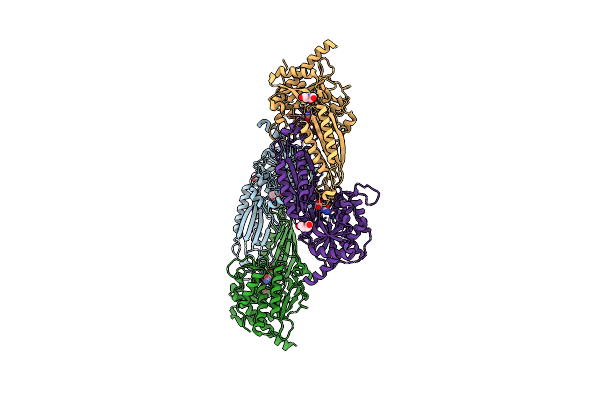
Crystal Structure Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With The Product Beta-Alanine
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, BAL, BCN |
 |
Crystal Structure Of Mutant E159A Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With Its Substrate N-Carbamyl-Beta- Alanine
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, URP, BCN |
 |
Crystal Structure Of Mutant R322A Of Beta-Alanine Synthase From Saccharomyces Kluyveri
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, URP, DTT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-01-04 Classification: APOPTOSIS Ligands: SO4 |
 |
Organism: Lachancea kluyveri
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2003-11-11 Classification: HYDROLASE Ligands: ZN, BIB |
 |
Crystal Structure Of Beta-Alanine Synthase From Saccharomyces Kluyveri (Selenomethionine Substituted Protein)
Organism: Lachancea kluyveri
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-11-11 Classification: HYDROLASE Ligands: ZN, DTT, TRS, BIB |

