Search Count: 12
 |
Crystal Structures Of The Cid1 Poly (U) Polymerase Reveal The Mechanism For Utp Selectivity
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-08-29 Classification: TRANSFERASE |
 |
Crystal Structures Of The Cid1 Poly (U) Polymerase Reveal The Mechanism For Utp Selectivity - Mgutp Bound
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-08-29 Classification: TRANSFERASE Ligands: UTP, MG |
 |
Crystal Structures Of The Cid1 Poly (U) Polymerase Reveal The Mechanism For Utp Selectivity - Cautp Bound
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-08-29 Classification: TRANSFERASE Ligands: UTP, CA, GOL |
 |
Crystal Structures Of The Cid1 Poly (U) Polymerase Reveal The Mechanism For Utp Selectivity - Mgctp Bound
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-08-29 Classification: TRANSFERASE Ligands: CTP, MG |
 |
Crystal Structures Of The Cid1 Poly (U) Polymerase Reveal The Mechanism For Utp Selectivity - Mggtp Bound
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-08-29 Classification: TRANSFERASE Ligands: GTP, MG |
 |
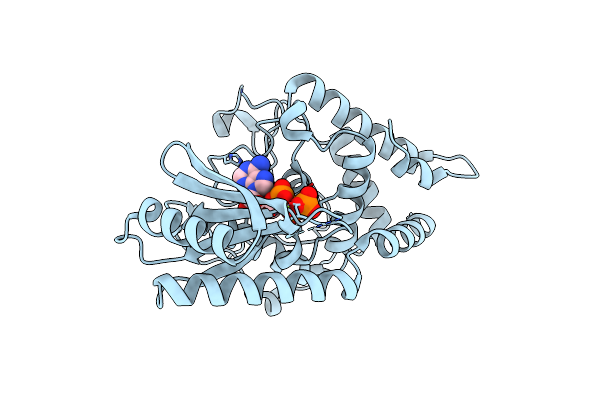
Crystal Structures Of The Cid1 Poly (U) Polymerase Reveal The Mechanism For Utp Selectivity - H336N Mutant Bound To Mgatp
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-08-29 Classification: TRANSFERASE Ligands: ATP, MG |
 |
Crystal Structures Of The Cid1 Poly (U) Polymerase Reveal The Mechanism For Utp Selectivity - Mg 3'-Datp Bound
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-08-29 Classification: TRANSFERASE Ligands: 3AT, MG |
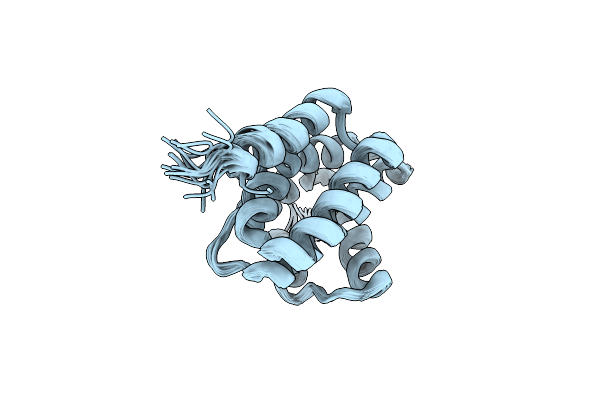 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2010-09-08 Classification: TRANSCRIPTION REGULATOR |
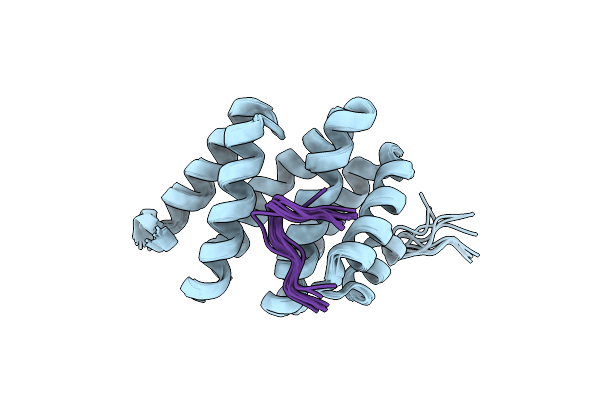 |
Solution Structure Of Rtt103 Ctd-Interacting Domain Bound To A Ser2 Phosphorylated Ctd Peptide
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2010-09-08 Classification: TRANSCRIPTION |
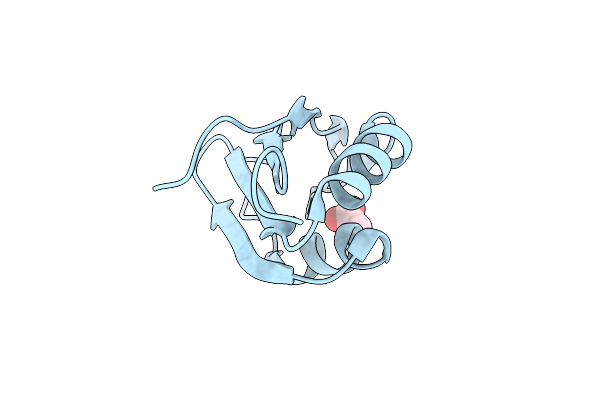 |
Structural Insights Into Cis Element Recognition Of Non- Polyadenylated Rnas By The Nab3-Rrm
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2010-09-08 Classification: RNA BINDING PROTEIN Ligands: ACT |
 |
Structural Insights Into Cis Element Recognition Of Non- Polyadenylated Rnas By The Nab3-Rrm
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-09-08 Classification: RNA BINDING PROTEIN/RNA Ligands: ACT |
 |
Nmr Structure Of Cfr (C-Terminal Fragment Of Computationally Designed Novel-Topology Protein Top7)
|

