Search Count: 41
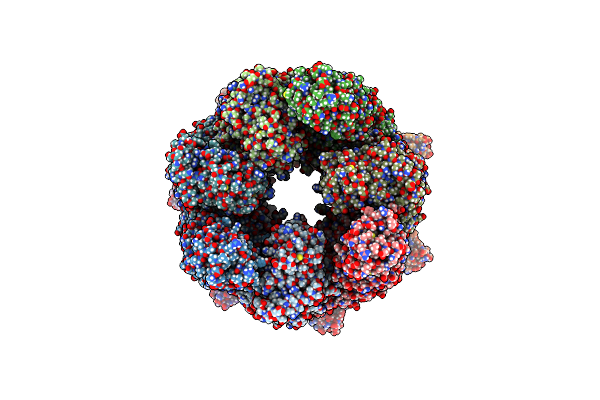 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: CHAPERONE Ligands: BEF, ADP, MG, K |
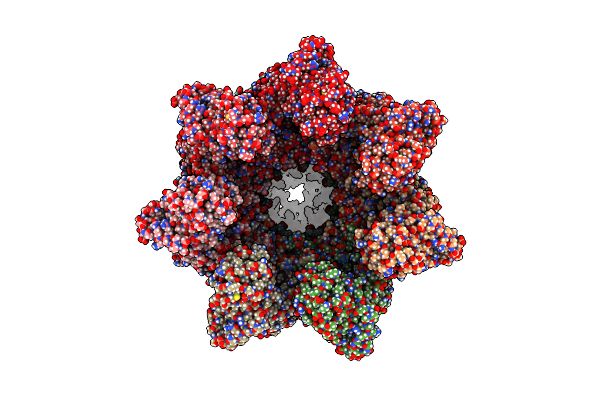 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: CHAPERONE Ligands: AF3, ADP, MG, K |
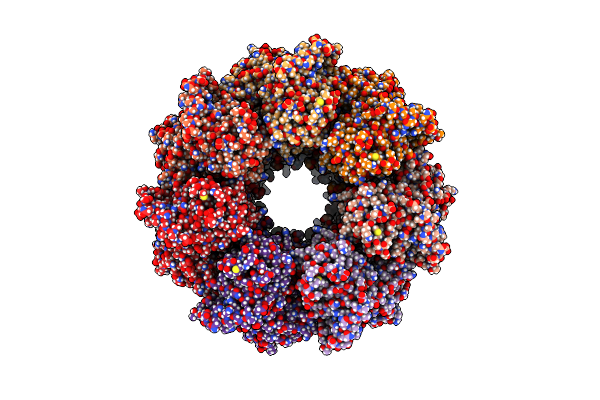 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: CHAPERONE |
 |
Organism: Pseudomonas monteilii
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
 |
Organism: Pseudomonas monteilii
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
 |
Organism: Pseudomonas monteilii
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
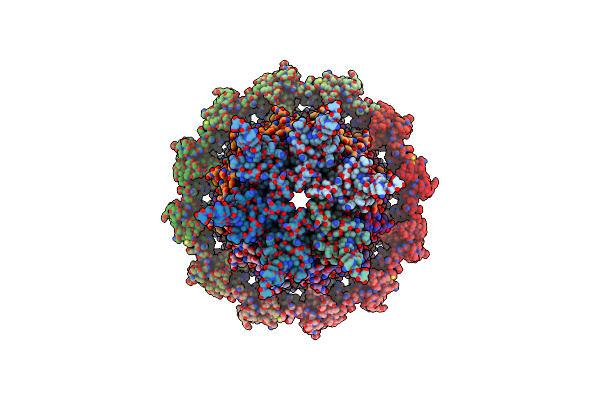 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: VIRAL PROTEIN |
 |
O-Layer Structure (Trwh/Virb7, Trwf/Virb9Ctd, Trwe/Virb10Ctd) Of The Outer Membrane Core Complex From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
 |
I-Layer Structure (Trwf/Virb9Ntd, Trwe/Virb10Ntd) Of The Outer Membrane Core Complex From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
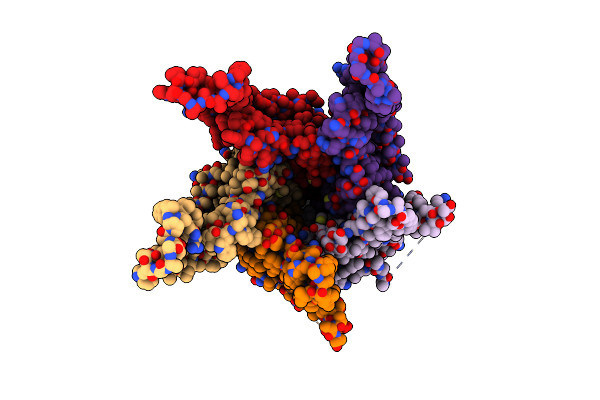 |
Stalk Complex Structure (Trwj/Virb5-Trwi/Virb6) From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
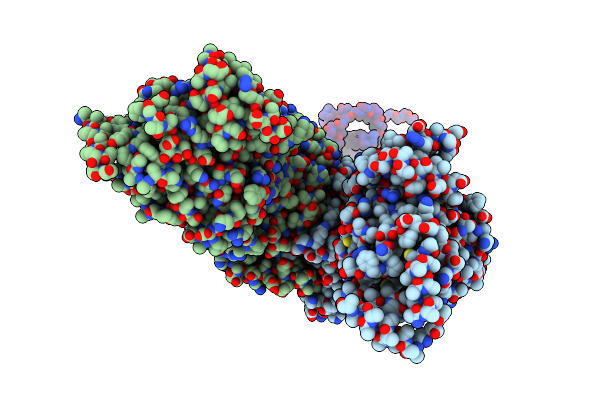 |
Hexameric Composite Model Of The Inner Membrane Complex (Imc) With The Arches From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
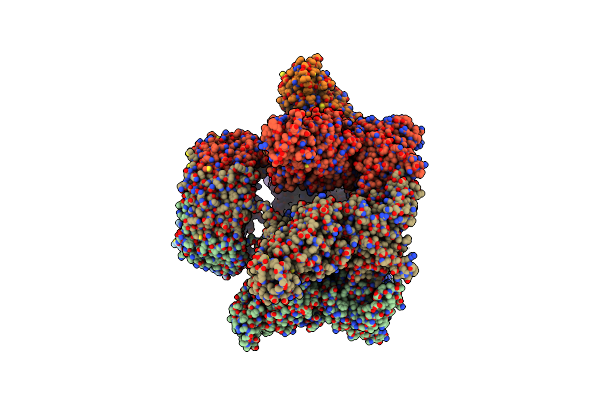 |
Trwk/Virb4Unbound Trimer Of Dimers Complex (With Hcp1) From The R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Salmonella dublin, Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
 |
Trwk/Virb4Unbound Dimer Complex From R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
 |
Inner Membrane Complex (Imc) Protomer Structure (Trwm/Virb3, Trwk/Virb4, Trwg/Virb8Tails) From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
 |
Arches Protomer (Trimer Of Trwg/Virb8Peri) Structure From The Fully-Assembled R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: IMMUNE SYSTEM Ligands: CA, NAG |
 |
L. Pneumophila Type Iv Coupling Complex (T4Cc) With Density For Doty N-Terminal And Middle Domains
Organism: legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
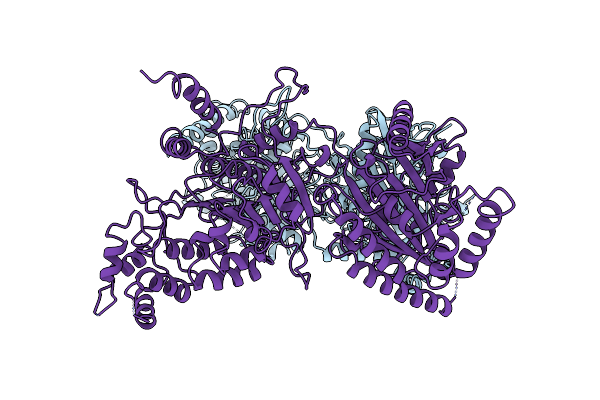
Cryo-Em Structure Of The Sulfolobus Acidocaldarius Rna Polymerase At 2.88 A
Organism: Sulfolobus acidocaldarius (strain atcc 33909 / dsm 639 / jcm 8929 / nbrc 15157 / ncimb 11770)
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: TRANSCRIPTION Ligands: MG, ZN, F3S |
 |
Cryo-Em Structure Of The Atv Rnap Inhibitory Protein (Rip) Bound To The Dna-Binding Channel Of The Host'S Rna Polymerase
Organism: Sulfolobus acidocaldarius dsm 639, Acidianus two-tailed virus
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: VIRAL PROTEIN Ligands: MG, ZN, F3S |
 |
Cryo-Em Structure Of The Cellular Negative Regulator Tfs4 Bound To The Archaeal Rna Polymerase
Organism: Sulfolobus acidocaldarius dsm 639, Saccharolobus solfataricus (strain atcc 35092 / dsm 1617 / jcm 11322 / p2)
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: TRANSCRIPTION Ligands: MG, ZN, F3S |

