Search Count: 60
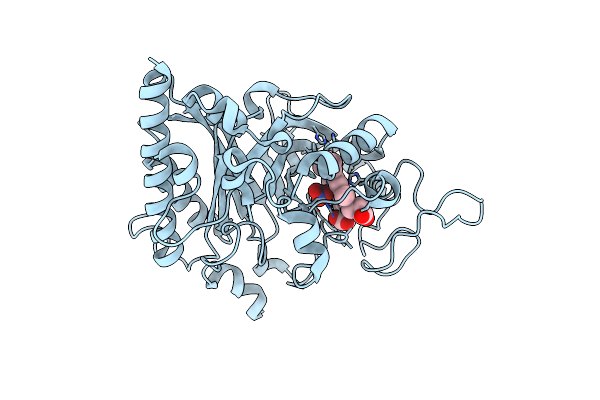 |
Organism: Thermobifida halotolerans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-02-28 Classification: OXIDOREDUCTASE Ligands: HEM, NO3 |
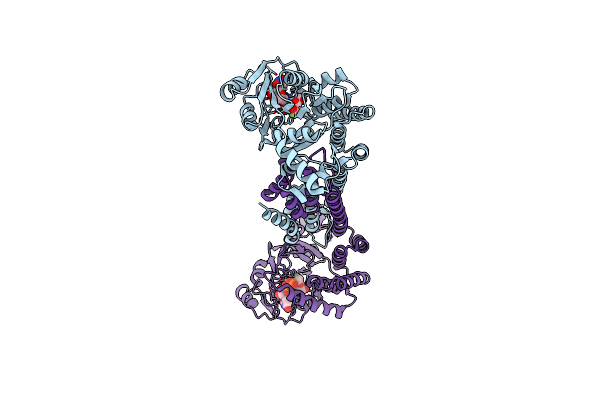 |
Organism: Bacillus thuringiensis serovar andalousiensis bgsc 4aw1
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-04-13 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, U2F |
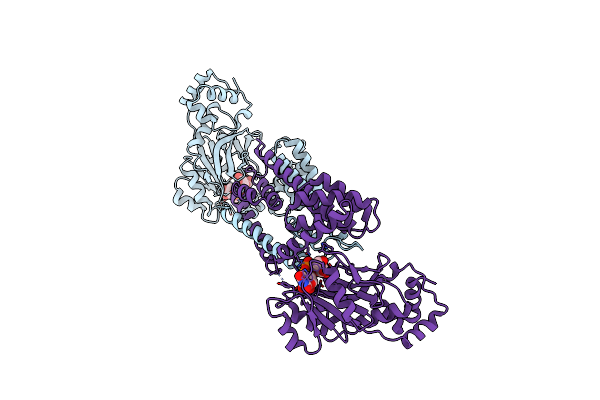 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-04-13 Classification: TRANSFERASE Ligands: UPG |
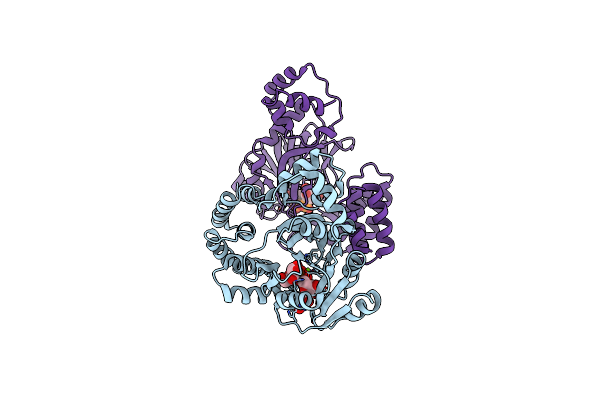 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-04-13 Classification: BIOSYNTHETIC PROTEIN Ligands: UPG, MG |
 |
Structure Of The M298F Mutant Of The Streptomyces Coelicolor Small Laccase T1 Copper Axial Ligand.
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-02-09 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Structure Of The M298L Mutant Of The Streptomyces Coelicolor Small Laccase T1 Copper Axial Ligand
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-02-09 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-02-02 Classification: TRANSFERASE Ligands: E8Q, CO3, SAH, SAM |
 |
Structure Of The Streptomyces Coelicolor Small Laccase - Cubic Crystal Form
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Structure Of The M198F M298F Double Mutant Of The Streptomyces Coelicolor Small Laccase T1 Copper Site
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: PG4, CU |
 |
First Crystal Structure Of Endo-Levanase Bt1760 From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-06-19 Classification: HYDROLASE Ligands: ZN, CL, MES, GOL |
 |
Endo-Levanase Bt1760 Mutant E221A From Bacteroides Thetaiotaomicron Complexed With Levantetraose
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-19 Classification: HYDROLASE Ligands: ZN, GOL |
 |
Crystal Structure Of Class Ii Lanthipeptide Synthetase Cylm In Complex With Amp
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-10-14 Classification: TRANSFERASE Ligands: ZN, AMP |
 |
Organism: Feline panleukopenia virus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-12-10 Classification: VIRUS Ligands: MG |
 |
Crystal Structure Of N-Methyl Transferase From Plasmodium Vivax Complexed With S-Adenosyl Methionine, Phosphate And Amodiaquine
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-10-01 Classification: transferase/transferase inhibitor Ligands: SAM, PO4, BME, CQA |
 |
Crystal Structure Of N-Methyl Transferase From Plasmodium Vivax Complexed With S-Adenosyl Methionine And Phosphate
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-02-19 Classification: TRANSFERASE Ligands: PO4, SAM, BME, EDO |
 |
Crystal Structure Of N-Methyl Transferase From Plasmodium Knowlesi Complexed With S-Adenosyl Methionine
Organism: Plasmodium knowlesi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-02-19 Classification: TRANSFERASE Ligands: SAM, BME |
 |
Crystal Structure Of N-Methyl Transferase (Pmt-2) From Caenorhabditis Elegant Complexed With S-Adenosyl Homocysteine And Phosphoethanolamine
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-01-15 Classification: TRANSFERASE Ligands: SAH, OPE, BME |
 |
Crystal Structure Of L-Fuconate Dehydratase From Silicibacter Sp. Tm1040 Liganded With Mg
Organism: Ruegeria sp. tm1040
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2013-12-04 Classification: ISOMERASE Ligands: GOL, MG, CO2 |
 |
Crystal Structure Of L-Fuconate Dehydratase From Silicibacter Sp. Tm1040 Liganded With Mg And D-Erythronohydroxamate
Organism: Ruegeria sp. tm1040
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2013-12-04 Classification: ISOMERASE Ligands: EHM, MG, GOL |
 |
Organism: Thermobifida cellulosilytica
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-09-11 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, GOL |

