Search Count: 20
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: BTB, GOL |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: EDO, PO4 |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: CL |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: C5J, CA |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-02-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: BTB, GOL, CL, C5S, PEG |
 |
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: FE, PEG |
 |
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: FE, EDO, PEG, CL |
 |
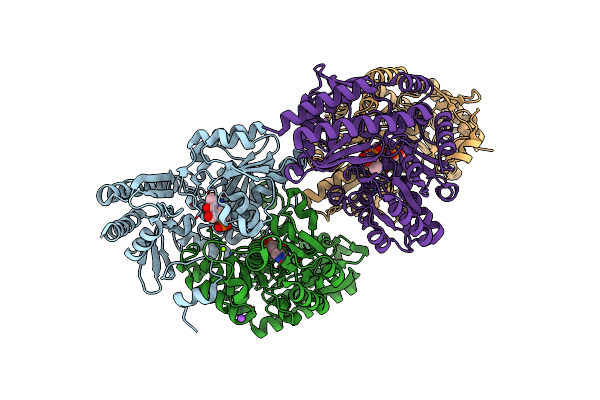
Structural And Functional Study Of Succinyl-Ornithine Transaminase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-01-16 Classification: TRANSFERASE Ligands: PLP, NA, MG |
 |
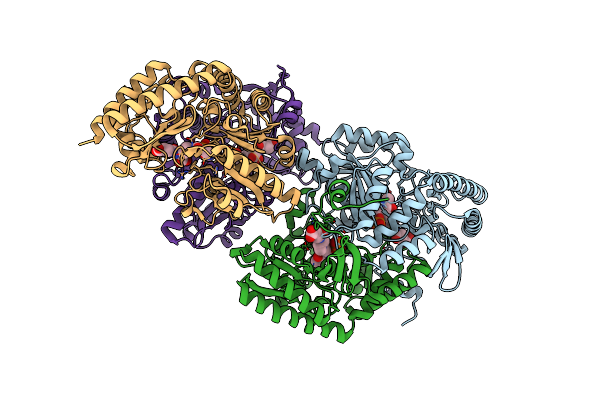
Structural And Functional Study Of Succinyl-Ornithine Transaminase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-01-16 Classification: TRANSFERASE Ligands: PLP, NA, MG |
 |
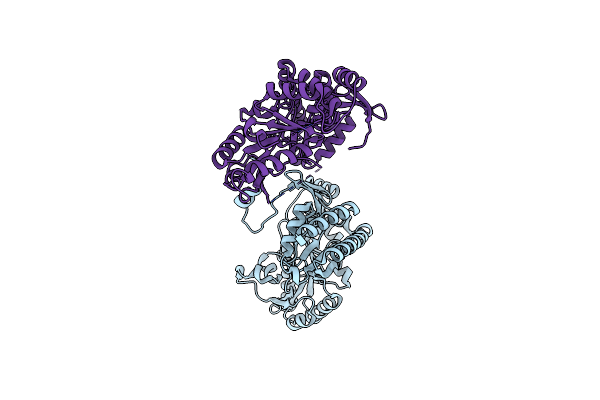
Structural And Functional Study Of Succinyl-Ornithine Transaminase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-01-16 Classification: TRANSFERASE Ligands: PLP, SUO |
 |
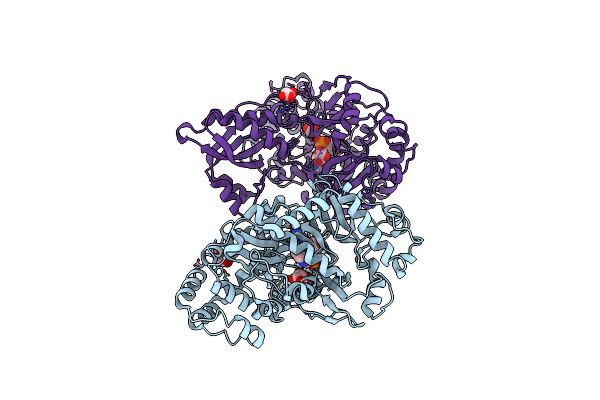
Structural And Functional Study Of Succinyl-Ornithine Transaminase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2013-01-16 Classification: TRANSFERASE |
 |
Crystal Structure Of An Indole-3-Acetic Acid Amido Synthase From Vitis Vinifera Involved In Auxin Homeostasis
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-12-19 Classification: SIGNALING PROTEIN Ligands: MLI, V1N |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-06-08 Classification: HYDROLASE Ligands: CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-06-08 Classification: HYDROLASE Ligands: CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2011-06-08 Classification: HYDROLASE Ligands: CL |
 |
Structural Basis For A New Mechanism Of Inhibition Of Hiv Integrase Identified By Fragment Screening And Structure Based Design
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, EDO, CL, ACY, IMV |
 |
Structural Basis For A New Mechanism Of Inhibition Of Hiv Integrase Identified By Fragment Screening And Structure Based Design
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, ACY, EDO, CIW |
 |
Structural Basis For A New Mechanism Of Inhibition Of Hiv Integrase Identified By Fragment Screening And Structure Based Design
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, CDQ, ACY |
 |
Structural Basis For A New Mechanism Of Inhibition Of Hiv Integrase Identified By Fragment Screening And Structure Based Design
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, CD9, EDO, ACY |
 |
Structural Basis For A New Mechanism Of Inhibition Of Hiv Integrase Identified By Fragment Screening And Structure Based Design
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, CBJ, ACY |

