Search Count: 23
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-06-08 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With The Dual Inhibitor Tw22
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-05-27 Classification: GENE REGULATION Ligands: EDO, P8N |
 |
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With The Dual Inhibitor Tw9
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2020-05-06 Classification: GENE REGULATION Ligands: EDO, P8T |
 |
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With The Dual Inhibitor Tw12
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2020-05-06 Classification: GENE REGULATION Ligands: EDO, P8Q |
 |
Crystal Structure Of Human Udp-N-Acetylglucosamine-Dolichyl-Phosphate N-Acetylglucosaminephosphotransferase (Dpagt1) (V264G Mutant) In Complex With Udp-Glcnac
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2018-07-25 Classification: TRANSFERASE Ligands: MG, UD1, UNL, P6L |
 |
Crystal Structure Of Human Udp-N-Acetylglucosamine-Dolichyl-Phosphate N-Acetylglucosaminephosphotransferase (Dpagt1) (V264G Mutant) In Complex With Tunicamycin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: P6L, 9LH, UNL |
 |
Crystal Structure Of Human Udp-N-Acetylglucosamine-Dolichyl-Phosphate N-Acetylglucosaminephosphotransferase (Dpagt1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: P6L |
 |
Crystal Structure Of Human Udp-N-Acetylglucosamine-Dolichyl-Phosphate N-Acetylglucosaminephosphotransferase (Dpagt1) (V264G Mutant)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-12-28 Classification: TRANSFERASE Ligands: UNL |
 |
 |
 |
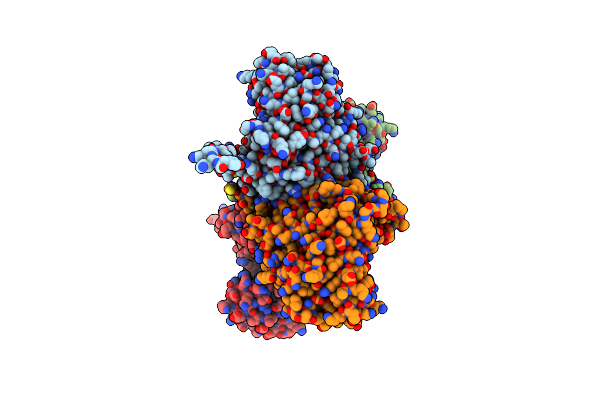 |
Crystal Structure Of Fructose-Bisphosphate Aldolase From Plasmodium Falciparum In Complex With Trap-Tail Determined At 2.7 Angstrom Resolution
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2007-04-17 Classification: LYASE |
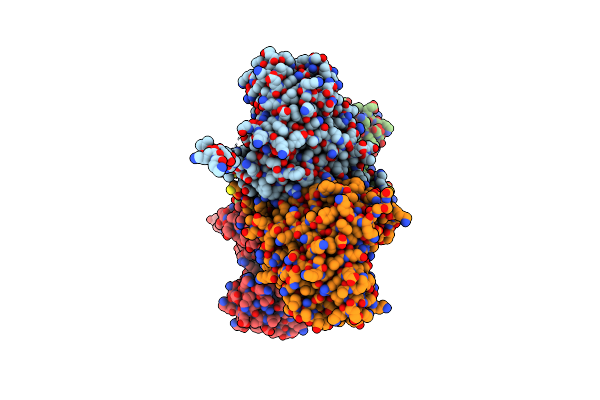 |
Crystal Structure Of Fructose-Bisphosphate Aldolase From Plasmodium Falciparum In Complex With Trap-Tail Determined At 2.4 Angstrom Resolution
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-04-17 Classification: LYASE |
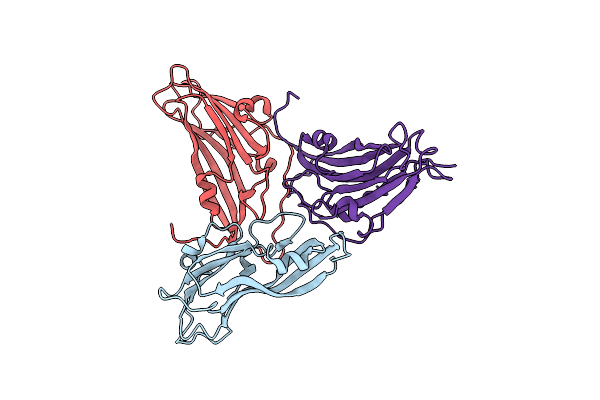 |
Organism: Turnip yellow mosaic virus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2006-02-21 Classification: VIRUS |
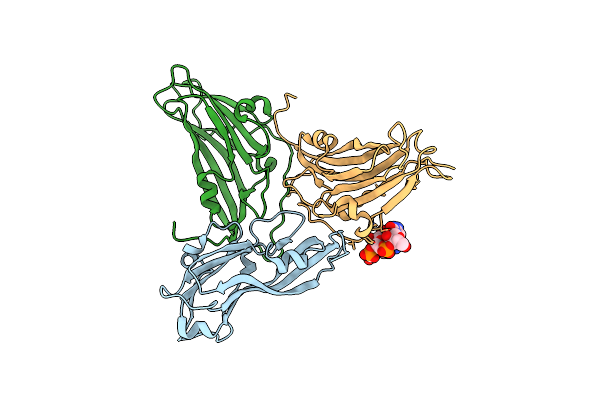 |
Organism: Turnip yellow mosaic virus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2006-02-21 Classification: Virus/RNA |
 |
Organism: Brome mosaic virus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2005-01-18 Classification: VIRUS |
 |
Organism: Tomato aspermy virus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2002-11-27 Classification: Virus/RNA Ligands: PO4, MG |
 |
Organism: Brome mosaic virus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2002-04-03 Classification: VIRUS Ligands: MG, P6G |
 |
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2001-01-10 Classification: OXYGEN STORAGE/TRANSPORT Ligands: HEM |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1997-03-12 Classification: OXYGEN TRANSPORT Ligands: ACT, SO4, HEM |

