Search Count: 142
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
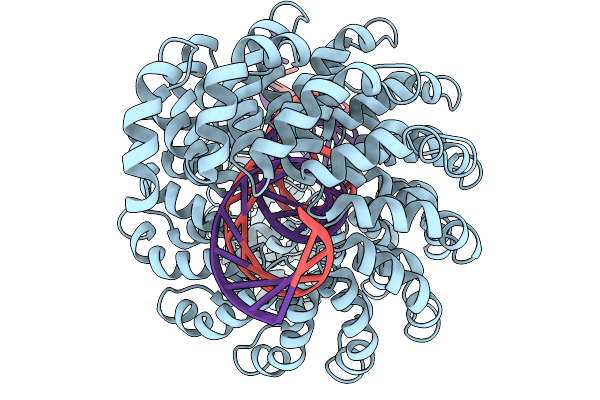 |
Organism: Burkholderia cenocepacia, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
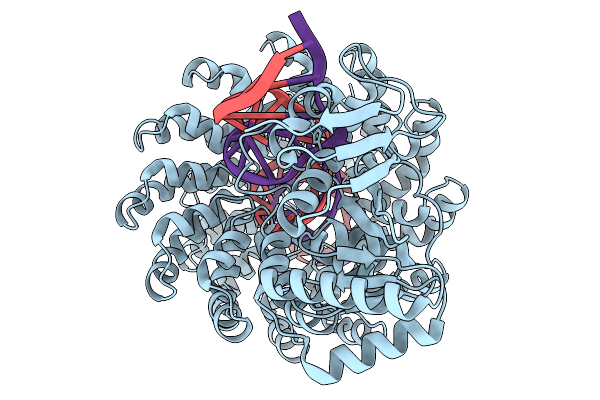 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
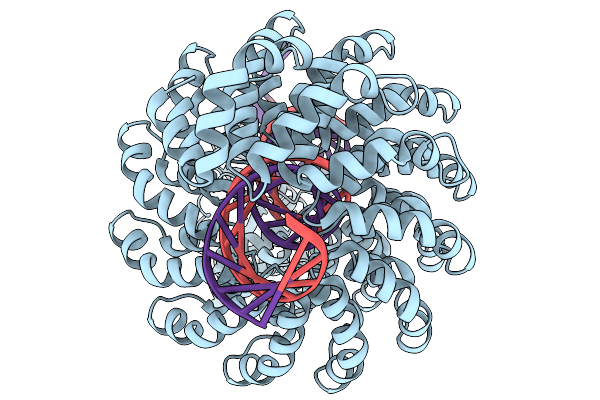 |
Organism: Xanthomonas, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
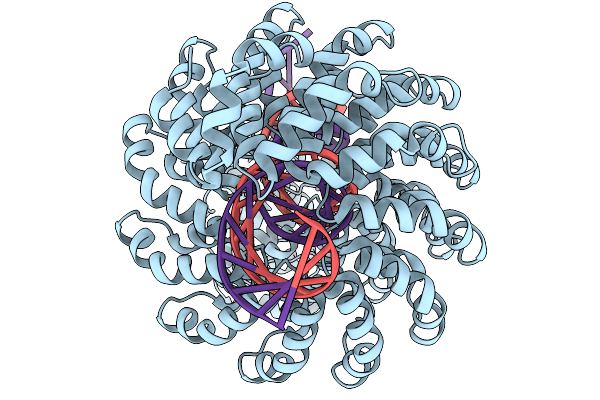 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
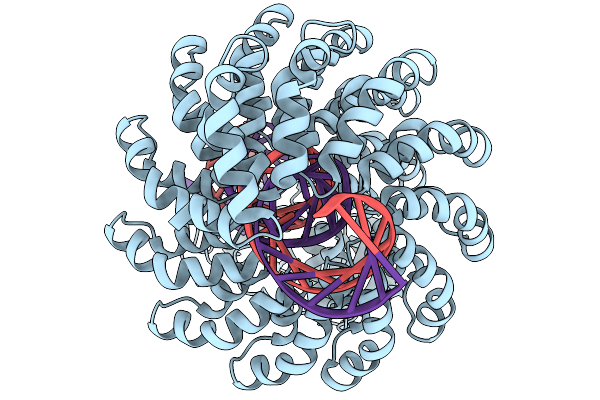 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
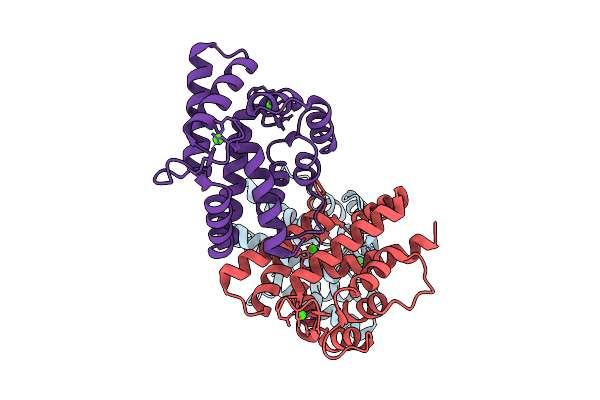 |
X-Ray Structure Of Sarcoplasmic Ca-Binding Protein (Scp), A Calcium Ion-Binding Protein From Pinctada Fucata
Organism: Pinctada fucata
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: CA |
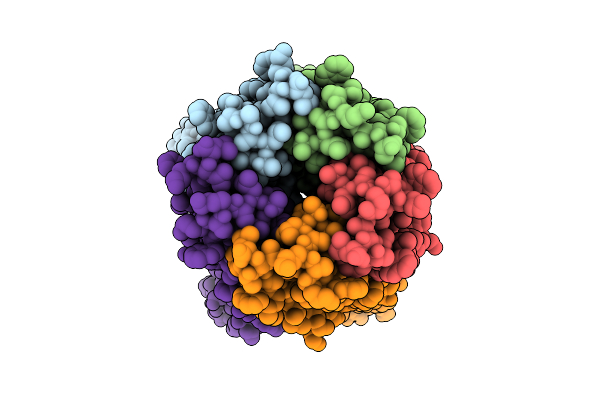 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DE NOVO PROTEIN Ligands: IOD |
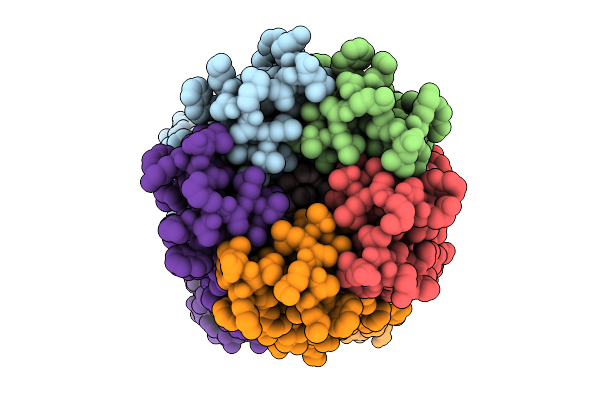 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN Ligands: LMT |
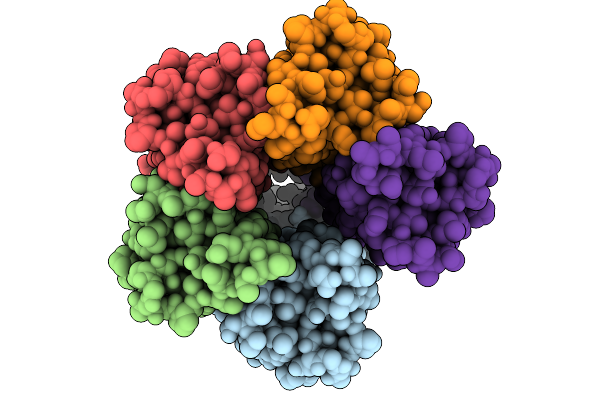 |
Cryo-Em Structure Of A De Novo Designed Voltage-Gated Anion Channel (Dvgac)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN |
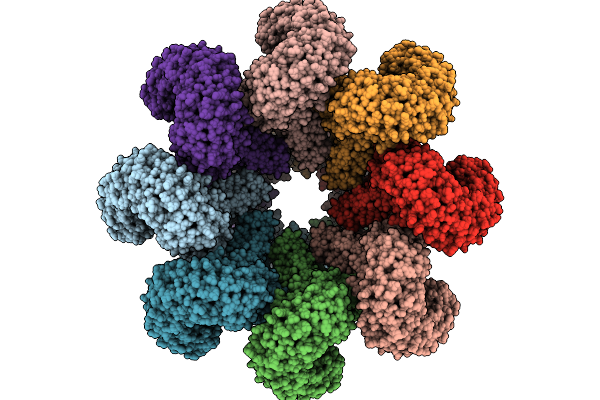 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat (N-To-N Arrangement)
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
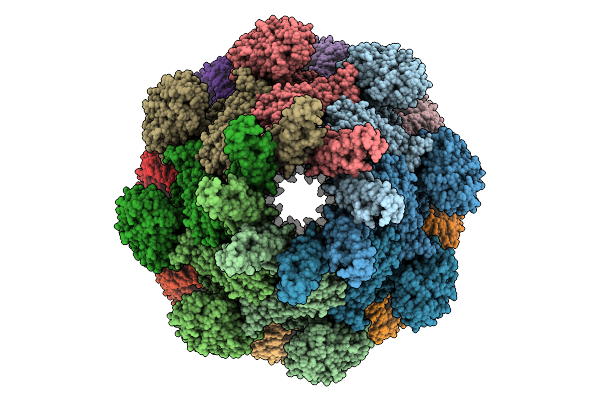 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat In 'Back-To-Back' Arrangement
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
Organism: Thermus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN |
 |
Improved Thermostability Of A Gh10 Xylanase Based On Its X-Ray Crystal Structure
Organism: Massilia sp. rbm26
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-06-11 Classification: CARBOHYDRATE |
 |
Cryo-Em Structure Of Apo Human Mitochondrial Pyruvate Carrier In The Ims-Open Conformation At Ph 8.0
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: PC8, CDL |
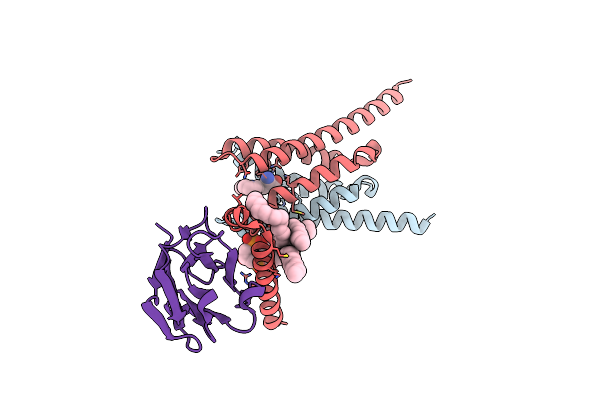 |
Cryo-Em Structure Of Human Mitochondrial Pyruvate Carrier In Complex With The Inhibitor Uk5099
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL, I2R |
 |
Cryo-Em Structure Of Human Mitochondrial Pyruvate Carrier In The Matrix-Facing Conformation At Ph 6.8
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL |
 |
Cryo-Em Structure Of Apo Human Mitochondrial Pyruvate Carrier In The Ims-Open Conformation At Ph 6.8
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL, PC8 |
 |
Cryo-Em Structure Of Human Mitochondrial Pyruvate Carrier In The Occluded Conformation At Ph 6.8
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CDL |

