Search Count: 152
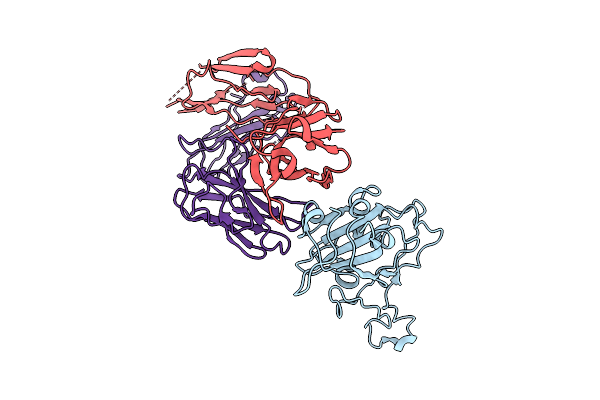 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
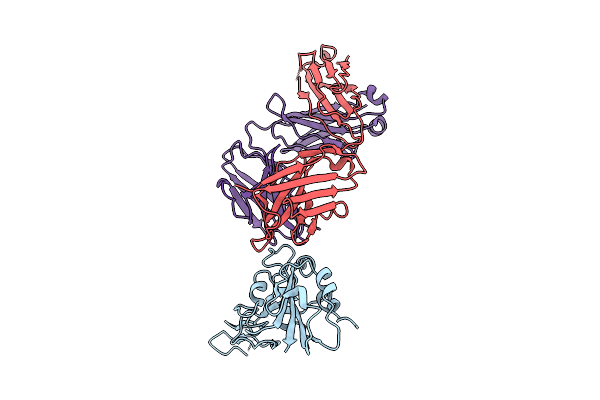
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
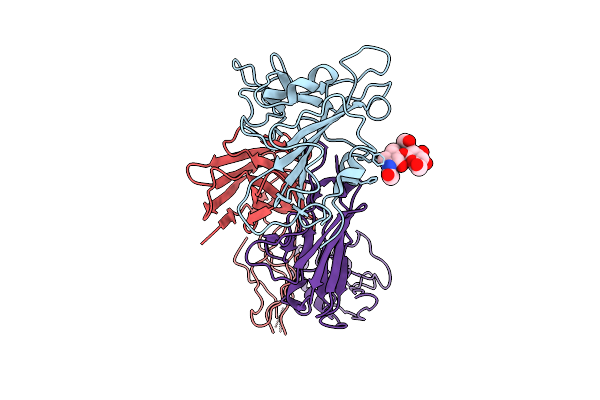
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Au-15330 In Complex With The Bromodomain Of Human Brm (Smarca2) And Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSCRIPTION Ligands: A1BNT, EDO, GOL |
 |
Crystal Structure Of Prt3789 In Complex With The Bromodomain Of Human Brg1 (Smarca4) And Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION Ligands: A1BB4, GOL, EDO, CL |
 |
Crystal Structure Of Prt3789 In Complex With The Bromodomain Of Human Brm (Smarca2) And Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION Ligands: A1BB4, CIT, ACT |
 |
Crystal Structure Of The Bromodomain Of Human Brm (Smarca2) In Complex With A Smarca-Bd Binder
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSCRIPTION Ligands: A1BB5, ZN |
 |
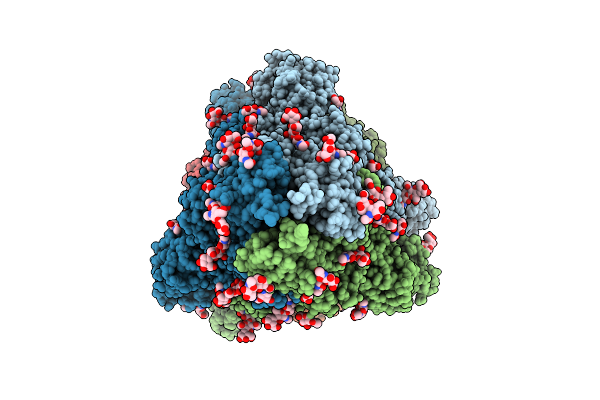
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat (N-To-N Arrangement)
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat In 'Back-To-Back' Arrangement
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
A Chimera Fadr Transcription Factor Containing Whth Of Rv0494 And The Part Other Than Whth Of Escherichia Coli Fadr
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSCRIPTION |
 |
Organism: Legionella pneumophila subsp. pneumophila (strain philadelphia 1 / atcc 33152 / dsm 7513)
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Coronavirus hku15
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2025-05-21 Classification: VIRAL PROTEIN |
 |
Organism: Coronavirus hku23
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2025-05-21 Classification: VIRAL PROTEIN Ligands: CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-05-07 Classification: DE NOVO PROTEIN Ligands: WUP, CL |
 |
Cryo-Em Structure Of Sars-Cov-2 D614G S With One Ace2 Receptor Binding (Rb1) In Prefusion Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |

