Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Brucella suis 1330
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: ADP, SO4, CL |
 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad (P1 Form2)
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: NAD, NA |
 |
Crystal Structure Of An Iole Protein From Brucella Melitensis (Hexagonal P Form)
Organism: Brucella melitensis biotype 1 (strain atcc 23456 / ccug 17765 / nctc 10094 / 16m)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: MN |
 |
Crystal Structure Of An Iole Protein From Brucella Melitensis (Orthorhombic P Form)
Organism: Brucella melitensis biotype 1 (strain atcc 23456 / ccug 17765 / nctc 10094 / 16m)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: MN |
 |
Crystal Structure Of A Iole Protein From Brucella Melitensis (Orthorhombic P Form 2)
Organism: Brucella melitensis biotype 1 (strain atcc 23456 / ccug 17765 / nctc 10094 / 16m)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: MN |
 |
Crystal Structure Of An Iole Protein From Brucella Melitensis (Cobalt Complex)
Organism: Brucella melitensis biotype 1 (strain atcc 23456 / ccug 17765 / nctc 10094 / 16m)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: CO |
 |
Crystal Structure Of 6,7-Dimethyl-8-Ribityllumazine Synthase From Bordetella Pertussis
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: CL, PG4, BCN |
 |
Crystal Structure Of 6,7-Dimethyl-8-Ribityllumazine Synthase From Bordetella Pertussis In Complex With 6,7-Dimethyl-8-(1'-D-Ribityl) Lumazine
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: PO4, CL, DLZ |
 |
Crystal Structure Of Nucleoside-Diphosphate Kinase Cryptosporidium Parvum (Apo, Hexamer)
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: EDO, CL |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Atp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: ATP, PEG, MG, PGE, GOL, ADP, PG4 |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Adp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: ADP, PEG, MG, PGE, GOL, PG4 |
 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Amp Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: AMP, PGE, MG, PEG, PG4, GOL |
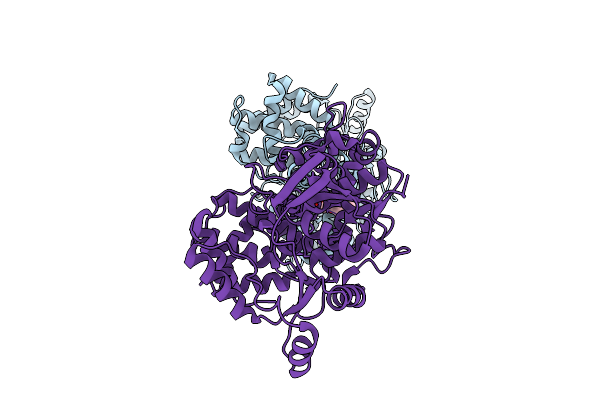 |
Crystal Structure Of Glutamate--Trna Ligase (Gltx) From Moraxella Catarrhalis (Apo)
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LIGASE Ligands: SO4, P6G, CL |
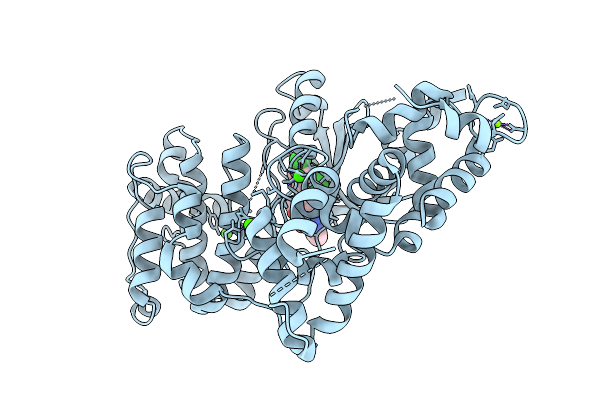 |
Crystal Structure Of Calcium-Dependent Protein Kinase 1 (Cdpk1) From Cryptosporidium Parvum In Complex With Inhibitor Win-3-115
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: CA, A1BLB, MG, CL |
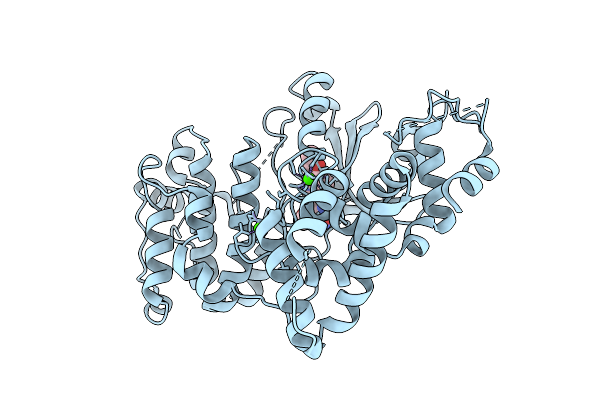 |
Crystal Structure Of Calcium-Dependent Protein Kinase 1 (Cdpk1) From Cryptosporidium Parvum In Complex With Inhibitor Bki-1606
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: A1BLC, CA |
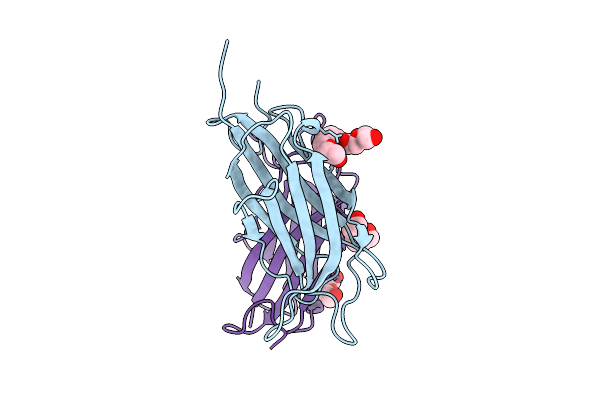 |
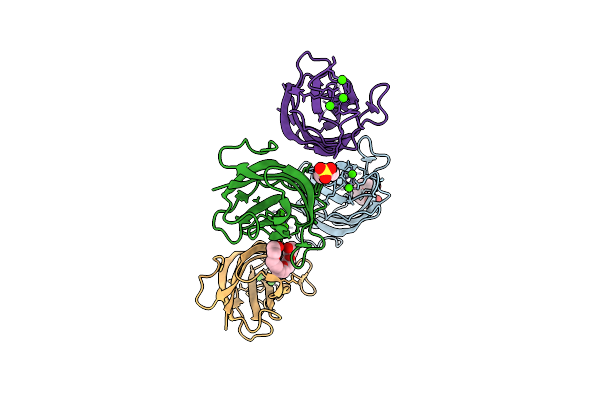
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: 1PE, CL, PEG, PGE |
 |
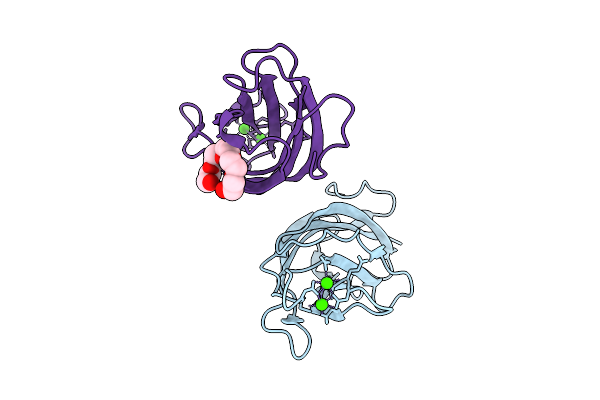
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: PG4, CA, MES, 1PE |
 |
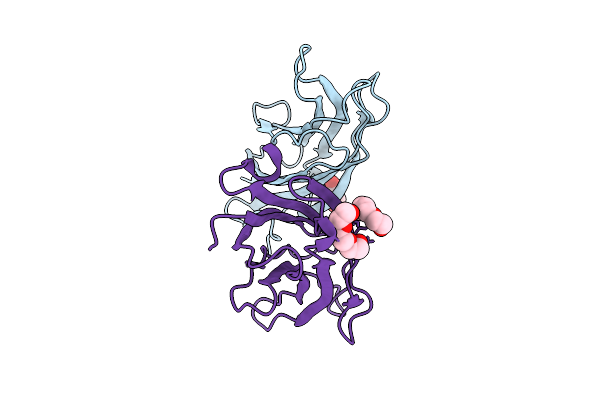
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (Orthorhombic P Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: CA, 1PE |
 |
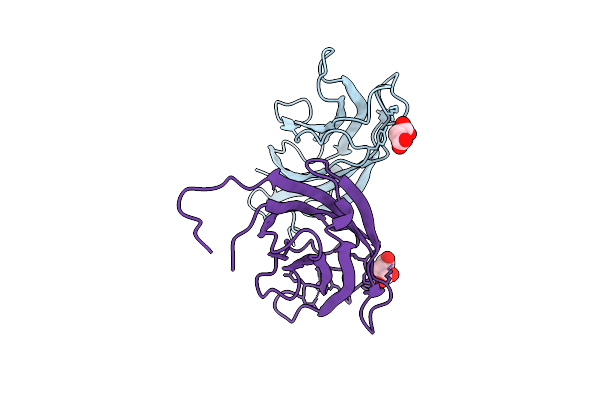
Crystal Structure Of A C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: GOL, PGE |
 |
Crystal Structure Of A Malonate Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: MLI, CL, NA |