Search Count: 137
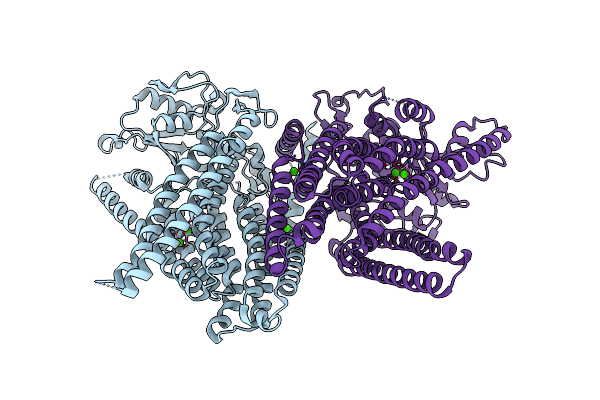 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-05-01 Classification: LIPID TRANSPORT Ligands: CA |
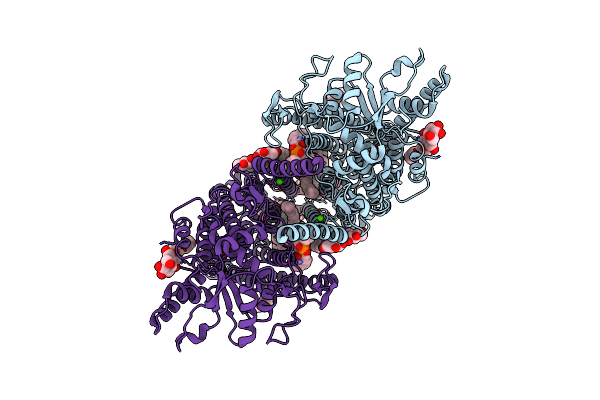 |
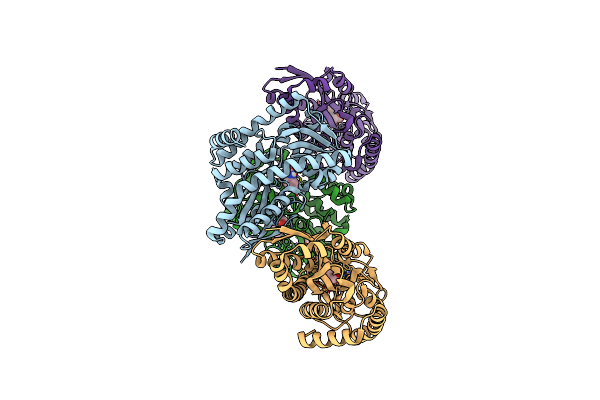
Cryoem Structure Of Calcium-Bound Human Tmem16K / Anoctamin 10 In Detergent (2Mm Ca2+, Closed Form)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-05-01 Classification: LIPID TRANSPORT Ligands: CA, PC1, UMQ |
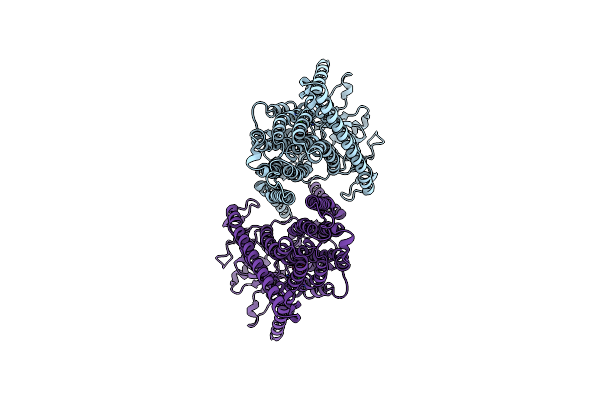 |
Cryoem Structure Of Calcium-Bound Human Tmem16K / Anoctamin 10 In Detergent (Low Ca2+, Closed Form)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-05-01 Classification: LIPID TRANSPORT Ligands: CA |
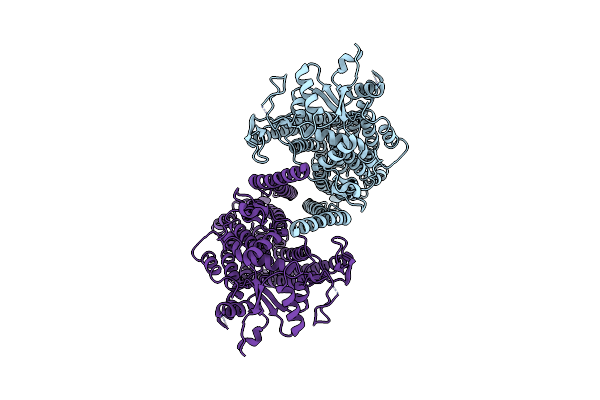 |
Cryoem Structure Of Calcium-Free Human Tmem16K / Anoctamin 10 In Detergent (Closed Form)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-05-01 Classification: LIPID TRANSPORT |
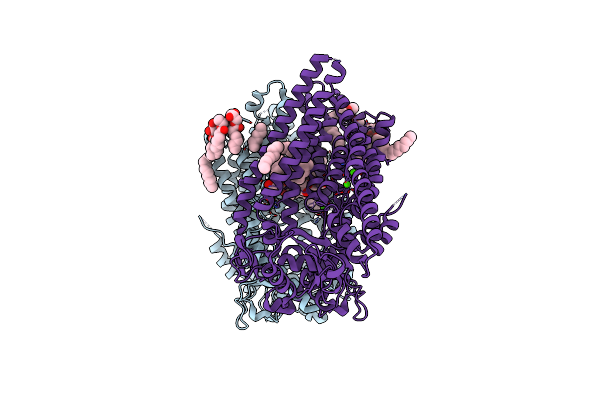 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-07-25 Classification: LIPID TRANSPORT Ligands: CA, 79M |
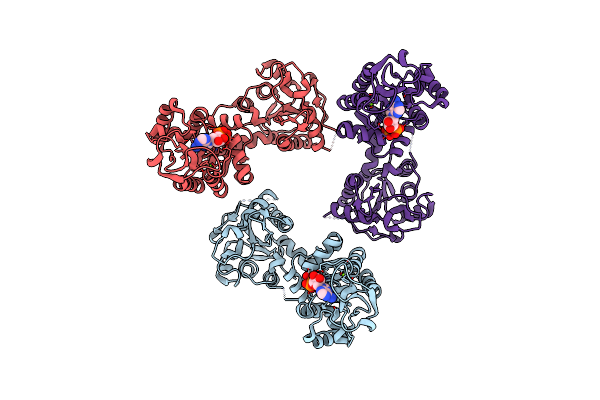 |
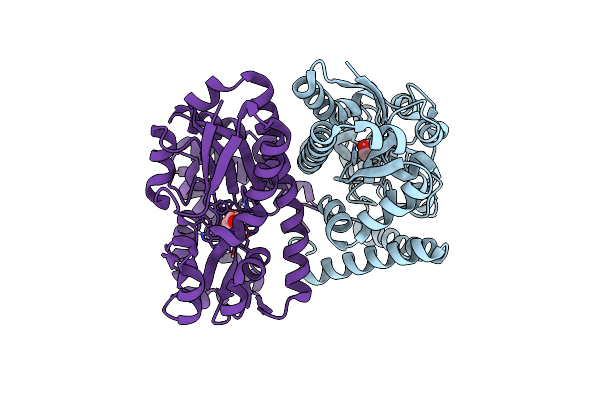
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2015-06-24 Classification: TRANSFERASE Ligands: MG, ADP |
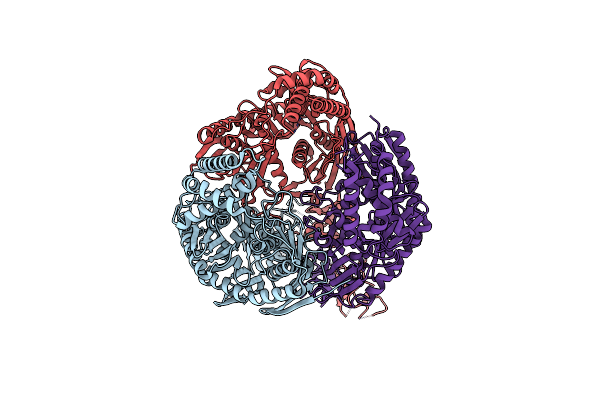 |
Structure Of The Polynucleotide Phosphorylase (Cbu_0852) From Coxiella Burnetii
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2015-06-17 Classification: TRANSFERASE Ligands: SO4 |
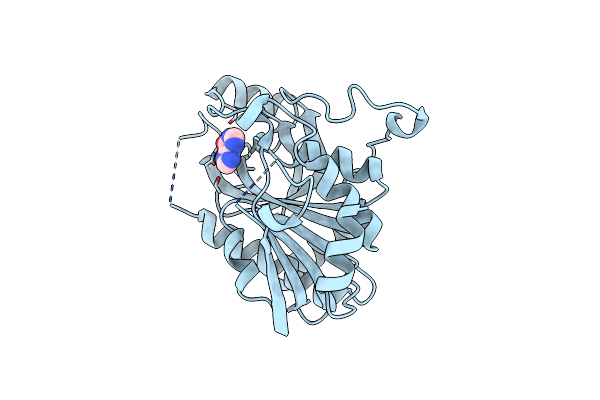 |
Crystal Structure Of A Purine Nucleoside Phosphorylase (Psi-Nysgrc-029736) From Agrobacterium Vitis
Organism: Agrobacterium vitis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-10-08 Classification: TRANSFERASE Ligands: HPA |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Chromohalobacter Salexigens Dsm 3043 (Csal_0678), Target Efi-501078, With Bound Ethanolamine
Organism: Chromohalobacter salexigens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-09-03 Classification: TRANSPORT PROTEIN Ligands: CL, ETA |
 |
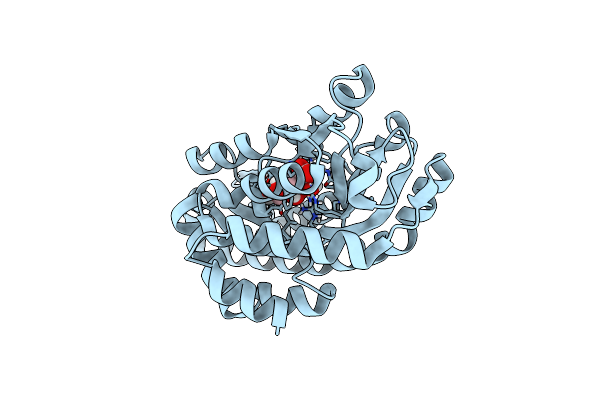
Crystal Structure Of A Putative Uridine Phosphorylase From Actinobacillus Succinogenes 130Z (Target Nysgrc-029667 )
Organism: Actinobacillus succinogenes
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-08-27 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Desulfovibrio Alaskensis G20 (Dde_0634, Target Efi-510120) With Bound 3-Indole Acetic Acid
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-06-18 Classification: SOLUTE-BINDING PROTEIN Ligands: IAC, EDO |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Desulfovibrio Alaskensis G20 (Dde_0634, Target Efi-510120) With Bound Indole Pyruvate
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-05-28 Classification: TRANSPORT PROTEIN Ligands: 3IO, EDO |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Rosenbacter Denitrificans Och 114 (Rd1_1052, Target Efi-510238) With Bound D-Mannonate
Organism: Roseobacter denitrificans
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-05-21 Classification: SOLUTE-BINDING PROTEIN Ligands: CS2 |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Polaromonas Sp Js666 (Bpro_0088, Target Efi-510167) Bound To D-Erythronate
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-05-14 Classification: SOLUTE-BINDING PROTEIN Ligands: EAX, MLA, FMT |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Polaromonas Sp Js666 (Bpro_1871, Target Efi-510164) Bound To D-Erythronate
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-05-14 Classification: SOLUTE-BINDING PROTEIN Ligands: EAX |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Rhodobacter Sphaeroides (Rsph17029_3620, Target Efi-510199), Apo Open Structure
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-05-14 Classification: SOLUTE-BINDING PROTEIN Ligands: IMD, EDO, ACT, ZN |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Colwellia Psychrerythraea (Cps_0129, Target Efi-510097) With Bound Calcium And Pyruvate
Organism: Colwellia psychrerythraea
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-05-14 Classification: SOLUTE-BINDING PROTEIN Ligands: CL, PYR, CA |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Roseobacter Denitrificans (Rd1_0742, Target Efi-510239) With Bound 3-Deoxy-D-Manno-Oct-2-Ulosonic Acid (Kdo)
Organism: Roseobacter denitrificans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-14 Classification: TRANSPORT PROTEIN Ligands: KDO, PGE, SO4 |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Sulfitobacter Sp. Nas-14.1 (Target Efi-510299) With Bound Beta-D-Galacturonate
Organism: Sulfitobacter sp. nas-14.1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-05-14 Classification: TRANSPORT PROTEIN Ligands: CL, GTR |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Fusobacterium Nucleatun (Fn1258, Target Efi-510120) With Bound Sn-Glycerol-3-Phosphate
Organism: Fusobacterium nucleatum subsp. nucleatum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-05-14 Classification: TRANSPORT PROTEIN Ligands: ZN, IMD, G3P |

