Search Count: 94
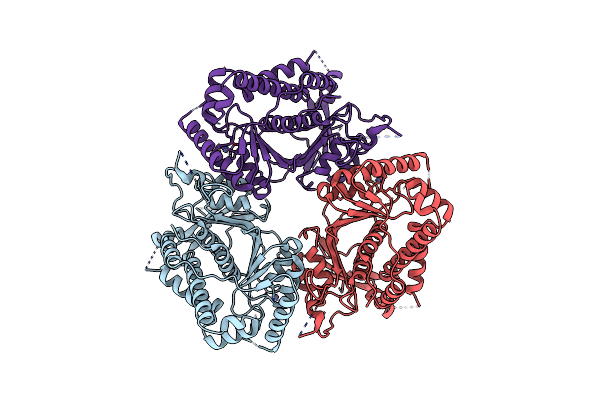 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Trimer Without A Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN |
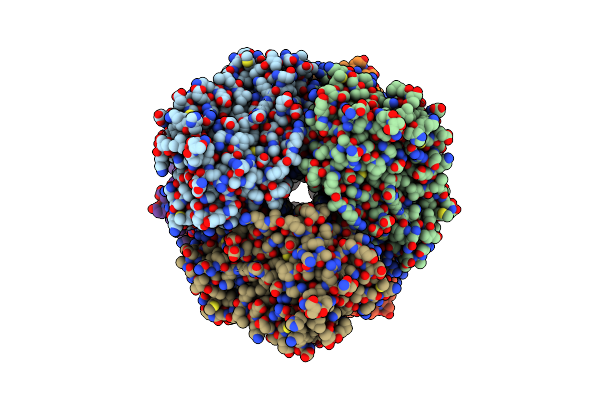 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer Without A Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN |
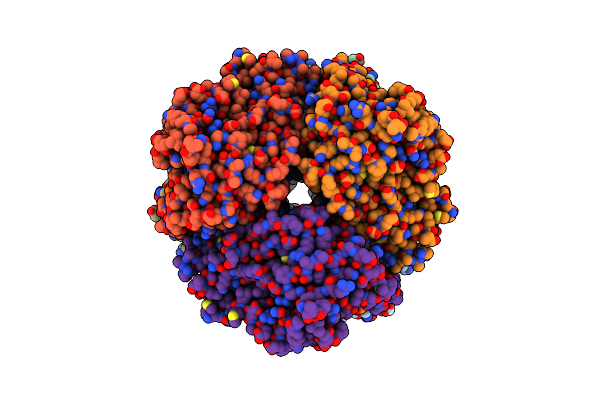 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer With Dctp Bound In The Active Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN, DCP |
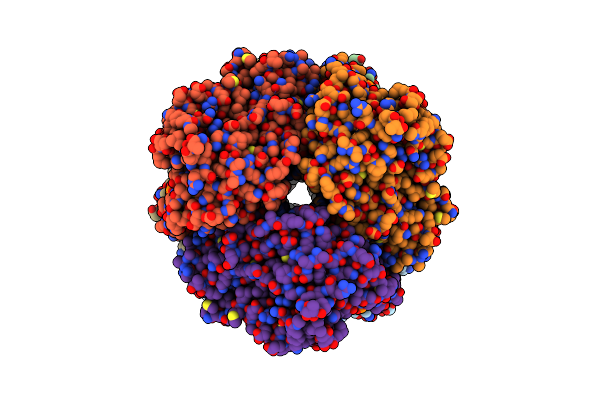 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer With 5Mdctp Bound In The Active Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN, A1I2I |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-09-25 Classification: DNA BINDING PROTEIN Ligands: XK6, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-09-25 Classification: DNA BINDING PROTEIN,HYDROLASE/INHIBITOR Ligands: Y9G, EDO, DMS, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-09-25 Classification: DNA BINDING PROTEIN,HYDROLASE/INHIBITOR Ligands: EDO, YA5, PEG, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-25 Classification: DNA BINDING PROTEIN,HYDROLASE/INHIBITOR Ligands: YCI, EDO, DMS, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-09-25 Classification: DNA BINDING PROTEIN,HYDROLASE/INHIBITOR Ligands: EDO, YE3, PEG, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-09-25 Classification: DNA BINDING PROTEIN Ligands: A1AIM, EDO, DMS |
 |
Crystal Structure Of Clostridioides Difficile Protein Tyrosine Phosphatase At Ph 8.5
Organism: Clostridioides difficile 6050
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-07-05 Classification: HYDROLASE |
 |
Crystal Structure Of Clostridioides Difficile Protein Tyrosine Phosphatase At Ph 7.5
Organism: Clostridioides difficile 6050
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2023-07-05 Classification: HYDROLASE Ligands: EPE, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: N8I, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: N7U, EDO, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: OYH, MPO, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: OYR, DMS, MPO, EDO |
 |
Crystal Structure Of Venezuelan Equine Encephalitis Alphavirus (Veev) Nonstructural Protein 2 (Nsp2) (K741A/K767A) Protease Domain
Organism: Venezuelan equine encephalitis virus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2023-03-15 Classification: VIRAL PROTEIN |
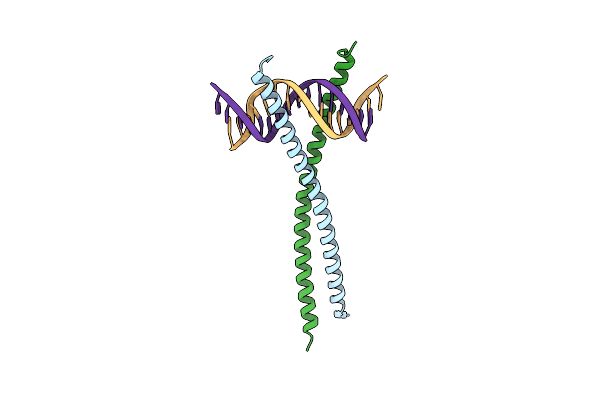 |
Structural Basis For Cell Type Specific Dna Binding Of C/Ebpbeta: The Case Of Cell Cycle Inhibitor P15Ink4B Promoter
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-11-23 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of Atp-Dependent Lon Protease From Bacillus Subtillis (Bslonba)
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: BO2, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2021-03-24 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: TGM, EDO |

