Search Count: 39
 |
Organism: Dragon grouper nervous necrosis virus
Method: SOLUTION NMR Release Date: 2024-10-30 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph8.0 (3.23A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph6.5 (2.82A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph5.0 (3.52A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virion At Ph6.5 (3.12A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Structural Basis Of Transcriptional Activation By The Ompr/Phob-Family Response Regulator Pmra
Organism: Escherichia coli bl21(de3), Klebsiella pneumoniae jm45, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2022-05-18 Classification: DNA BINDING PROTEIN |
 |
Organism: Staphylococcus aureus, Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-18 Classification: DNA BINDING PROTEIN/DNA Ligands: MG |
 |
Organism: Staphylococcus aureus (strain mu50 / atcc 700699)
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2022-05-18 Classification: DNA BINDING PROTEIN Ligands: BEF, MG |
 |
Crystal Structure Of C-Terminal Dna-Binding Domain Of Escherichia Coli Ompr
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.56 Å Release Date: 2020-12-23 Classification: DNA BINDING PROTEIN |
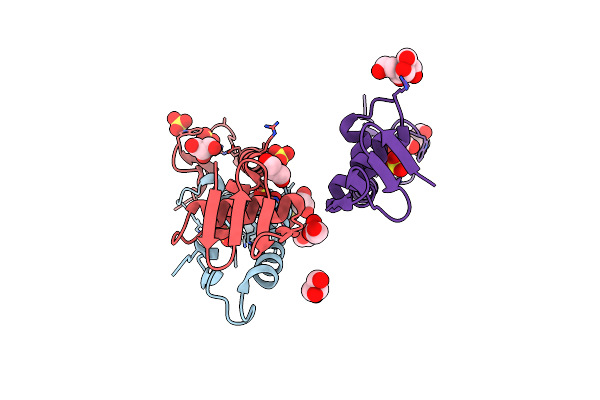 |
Crystal Structure Of C-Terminal Dna-Binding Domain Of Escherichia Coli Ompr As A Domain-Swapped Dimer
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2020-12-23 Classification: DNA BINDING PROTEIN Ligands: BTB, SO4, GOL |
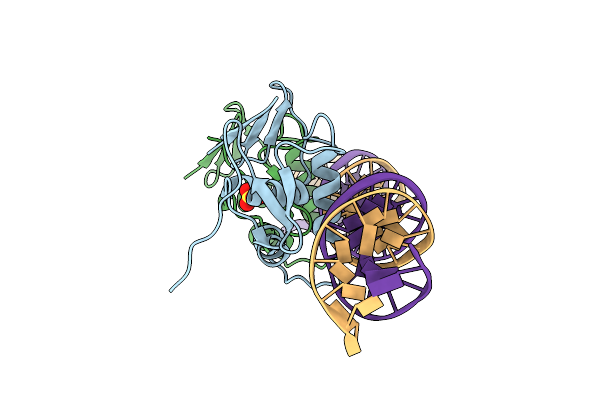 |
Crystal Structure Of C-Terminal Dna-Binding Domain Of Escherichia Coli Ompr In Complex With F1-Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2020-12-23 Classification: DNA BINDING PROTEIN Ligands: SO4 |
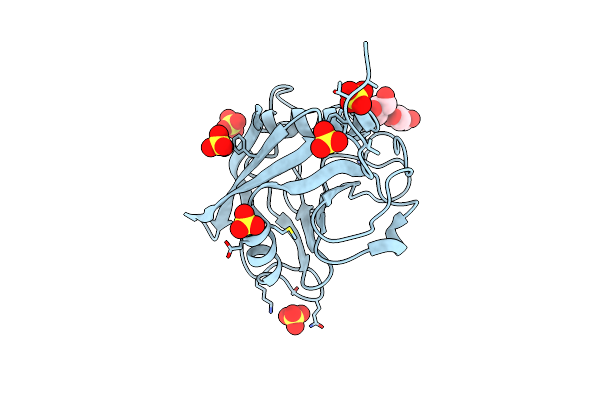 |
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2020-09-09 Classification: ISOMERASE Ligands: SO4, GOL |
 |
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-09-09 Classification: ISOMERASE |
 |
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-09-09 Classification: ISOMERASE Ligands: PO4, GOL |
 |
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2020-09-09 Classification: ISOMERASE |
 |
Organism: Pholiota squarrosa
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2020-03-04 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of A Mini Fungal Lectin, Phosl In Complex With Core-Fucosylated Chitobiose
Organism: Pholiota squarrosa
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2020-03-04 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
Crystal Structure Of Klebsiella Pneumoniae Sigma4 Of Sigmas Fusing With The Rna Polymerase Beta-Flap-Tip-Helix In Complex With -35 Element Dna
Organism: Escherichia coli, Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:3.75 Å Release Date: 2019-09-11 Classification: TRANSCRIPTION |
 |
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2018-07-18 Classification: ISOMERASE |

