Search Count: 64
 |
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: HEM, IMD, NA, CL |
 |
Organism: Plasmodium falciparum, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-03-26 Classification: PEPTIDE BINDING PROTEIN Ligands: AGS, PE4 |
 |
Organism: Coxsackievirus a6
Method: ELECTRON MICROSCOPY Resolution:3.15 Å Release Date: 2024-10-16 Classification: VIRUS LIKE PARTICLE |
 |
Structure Of The Non-Canonical Ctlh E3 Substrate Receptor Wdr26 Bound To Ypel5
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: LIGASE Ligands: ZN |
 |
Structure Of The Non-Canonical Ctlh E3 Substrate Receptor Wdr26 Bound To Nmnat1 Substrate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: LIGASE Ligands: NMN, ZN |
 |
Organism: Danio rerio
Method: SOLUTION NMR Release Date: 2024-05-08 Classification: METAL BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: IMMUNE SYSTEM Ligands: GDP, AF3, MG |
 |
Catalytic Module Of Human Ctlh E3 Ligase Bound To Multiphosphorylated Ube2H~Ubiquitin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIGASE Ligands: ZN |
 |
Catalytic Module Of Yeast Gid E3 Ligase Bound To Multiphosphorylated Ubc8~Ubiquitin
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIGASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-03-08 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-03-08 Classification: TRANSFERASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2023-02-08 Classification: LIGASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-02-08 Classification: LIGASE Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: LIGASE |
 |
Crystal Structure Of Sars-Cov-2 Spike Protein N-Terminal Domain In Complex With Biliverdin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-04-28 Classification: VIRAL PROTEIN Ligands: BLA, NAG, PG4, PEG, PGE, 1PE |
 |
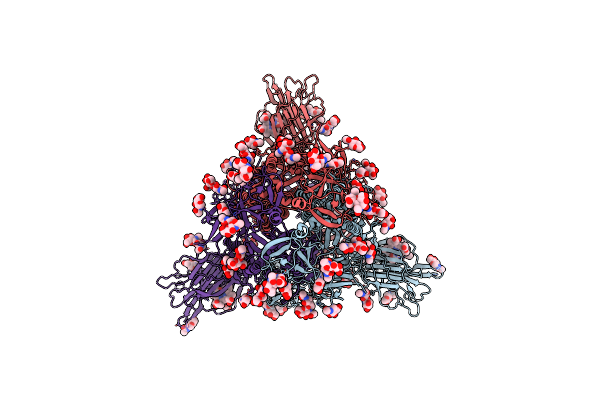
Trimeric Sars-Cov-2 Spike Ectodomain In Complex With Biliverdin (Closed Conformation)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: VIRAL PROTEIN Ligands: BLA, NAG |
 |
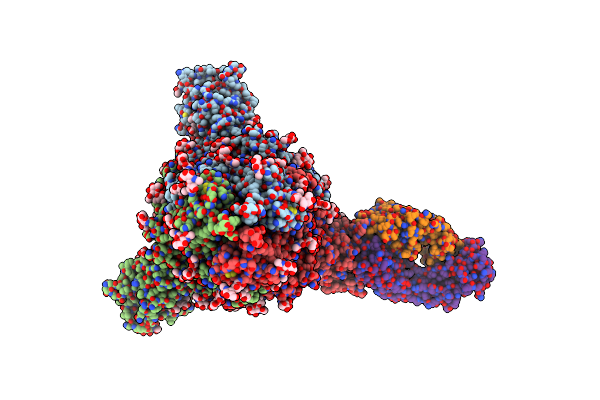
Trimeric Sars-Cov-2 Spike Ectodomain In Complex With Biliverdin (One Rbd Erect)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: VIRAL PROTEIN Ligands: BLA, NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: VIRAL PROTEIN Ligands: BLA, NAG |
 |
Crystal Structure Of Full-Length Cbnr Complexed With The Target Dna Complex
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2021-03-10 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of Human Methionine Aminopeptidase-2 In Complex With An Inhibitor Gw557358X (Compound 9)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-23 Classification: HYDROLASE Ligands: PO4, QVK, MN |

