Search Count: 65
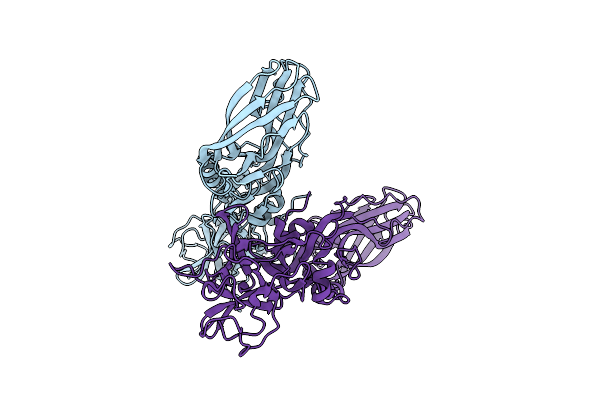 |
Structure Of The Lysinibacillus Sphaericus Tpp49Aa1 Pesticidal Protein At Ph 3
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-11-01 Classification: TOXIN |
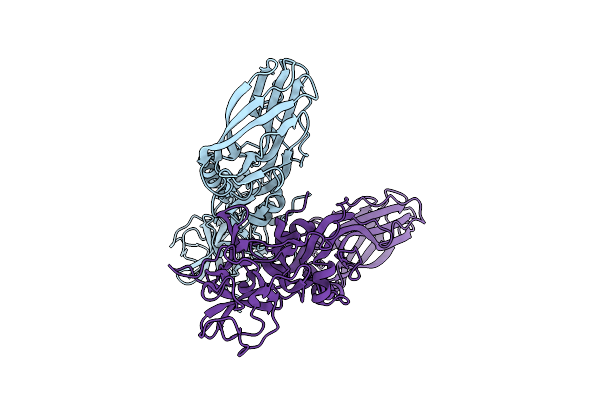 |
Structure Of The Lysinibacillus Sphaericus Tpp49Aa1 Pesticidal Protein At Ph 7
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-11-01 Classification: TOXIN |
 |
Structure Of The Lysinibacillus Sphaericus Tpp49Aa1 Pesticidal Protein At Ph 11
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-11-01 Classification: TOXIN |
 |
Organism: Oscillatoria acuminata pcc 6304
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-11-01 Classification: LYASE Ligands: FMN, MG |
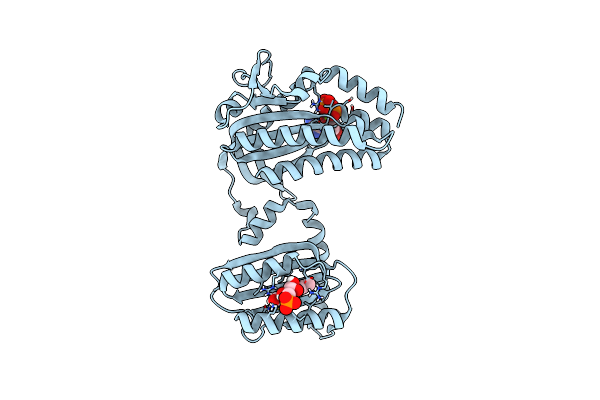 |
Cryogenic Crystal Structure Of The Photoactivated Adenylate Cyclase Oapac With Atp Bound
Organism: Oscillatoria acuminata pcc 6304
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-11-01 Classification: LYASE Ligands: FMN, ATP, MG |
 |
Cryogenic Crystal Structure Of The Photoactivated Adenylate Cyclase Oapac After 5 Seconds Of Blue Light Illumination
Organism: Oscillatoria acuminata pcc 6304
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-01 Classification: LYASE Ligands: FMN, ATP, MG |
 |
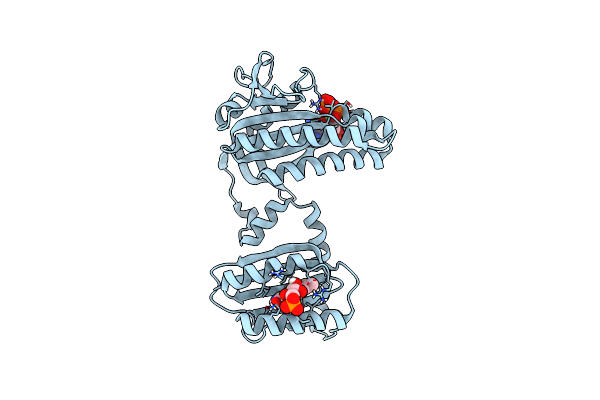
Room Temperature Crystal Structure Of The Photoactivated Adenylate Cyclase Oapac With Atp Bound
Organism: Oscillatoria acuminata pcc 6304
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-11-01 Classification: LYASE Ligands: FMN, ATP, MG |
 |
Room Temperature Crystal Structure Of The Photoactivated Adenylate Cyclase Oapac After Blue Light Excitation At 1.8 Us Delay
Organism: Oscillatoria acuminata pcc 6304
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: LYASE Ligands: FMN, ATP, MG |
 |
Room Temperature Crystal Structure Of The Photoactivated Adenylate Cyclase Oapac After Blue Light Excitation At 2.3 Us Delay
Organism: Oscillatoria acuminata pcc 6304
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: LYASE Ligands: FMN, ATP, MG |
 |
The Structure Of Natural Crystals Of The Lysinibacillus Sphaericus Tpp49Aa1 Pesticidal Protein Elucidated Using Serial Femtosecond Crystallography At An X-Ray Free Electron Laser
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-17 Classification: TOXIN |
 |
Crystal Structure Of Sars-Cov-2 Main Protease In Orthorhombic Space Group P212121
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: CL, DMS, MLI |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-01-25 Classification: VIRAL PROTEIN Ligands: DMS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-01-18 Classification: VIRAL PROTEIN Ligands: CL |
 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(2-Pyrrolidyl)-3,4,5-Trihydroxybenzoylhydrazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-03-23 Classification: HYDROLASE Ligands: DZI, GOL, ZN, CL, PO4 |
 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(3,5-Dimethoxy-4-Hydroxybenzyliden)Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: A5I, GOL, ZN, PO4, CL |
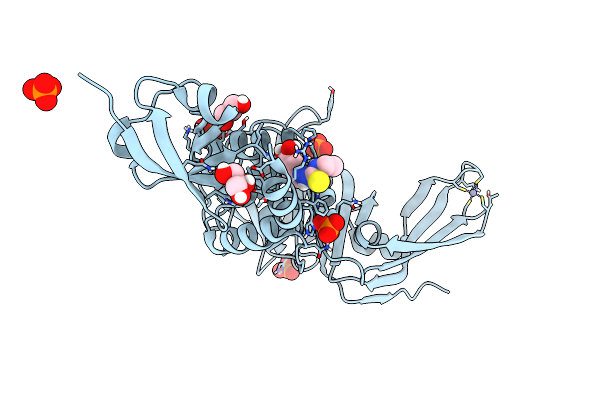 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(3,4-Dihydroxybenzylidene)-Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: A6Q, GOL, ZN, PO4, CL |
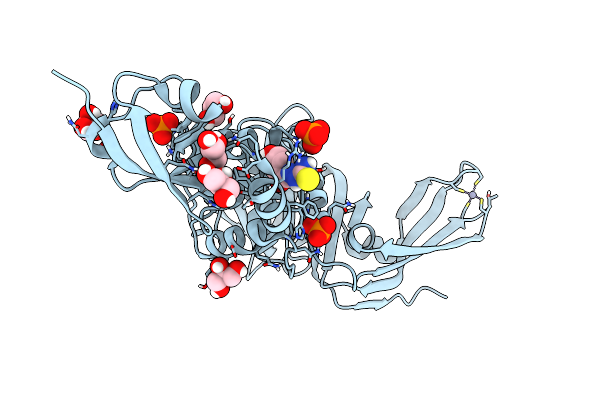 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(2,4-Dihydroxybenzylidene)-Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: GOL, A4O, ZN, PO4, CL |
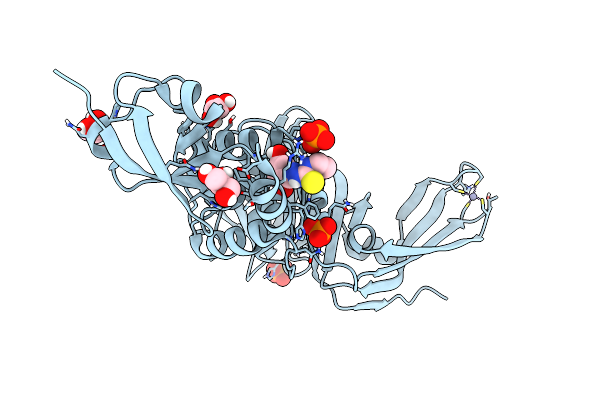 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(2,5-Dihydroxybenzylidene)-Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: A7L, GOL, ZN, PO4, CL |
 |
Structure Of Sars-Cov-2 Papain-Like Protease Bound To N-(3-Methoxy-4-Hydroxy-Acetophenone)Thiosemicarbazone
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-03-16 Classification: HYDROLASE Ligands: A3X, GOL, ZN, CL, PO4 |
 |
Structure Of Sars-Cov-2 Papain-Like Protease Plpro In Complex With 4-(2-Hydroxyethyl)Phenol
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-05-12 Classification: HYDROLASE Ligands: ZN, CL, YRL, PO4, GOL |

