Search Count: 33
 |
Organism: Pseudomonas fluorescens pf0-1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-04-09 Classification: HYDROLASE |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-04-09 Classification: HYDROLASE |
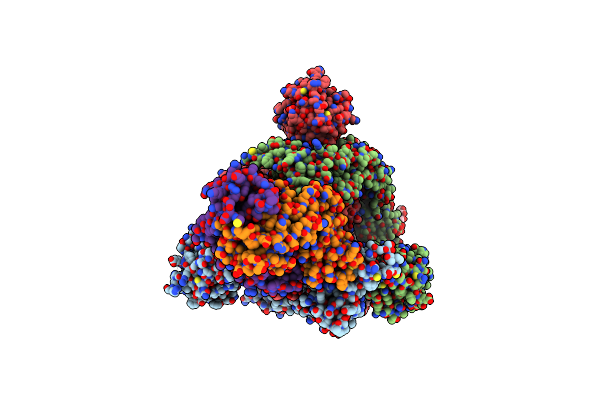 |
Sars-Cov-2 Spike Glycoprotein Trimer Complexed With Fab Fragment Of Anti-Rbd Antibody E7
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Sars-Cov-2 Spike Glycoprotein Trimer Complexed With Fab Fragment Of Anti-Rbd Antibody E7 (Focused Refinement On Fab-Rbd Interface)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: VIRAL PROTEIN |
 |
Gcf1P, Multimerizes And Bridges The Mitochondrial Dna From Candida Albicans By A Specific Mechanism.
Organism: Candida albicans, Synthetic construct
Method: X-RAY DIFFRACTION, SOLUTION SCATTERING Resolution:2.90 Å Release Date: 2023-06-14 Classification: DNA BINDING PROTEIN Ligands: GOL |
 |
Crystal Structure Of S/T Protein Kinase Pkng From Mycobacterium Tuberculosis In Complex With Inhibitor L2W
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: FE, 8ZC, NA |
 |
Nmr Structure Of The Zeta-Subunit Of The F1F0-Atpase From Sinorhizobium Meliloti
Organism: Sinorhizobium meliloti 1021
Method: SOLUTION NMR Release Date: 2021-10-13 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 6-[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]-N-(4-Hydroxy-3-Methoxybenzyl)Hexanamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8UH, NAG, PEG, CL |
 |
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 7-[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]-N-(4-Hydroxy-3-Methoxybenzyl)Heptanamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8UK |
 |
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 8-[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]-N-(4-Hydroxy-3-Methoxybenzyl)Octanamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: PEG, NAG, 8UQ, CL |
 |
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 4-{[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]Methyl}-N-(4-Hydroxy-3-Methoxybenzyl)Benzamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8UE |
 |
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With (E)-10-[(3-Chloro-6,7,10,11-Tetrahydro-9-Methyl-7,11-Methanocycloocta[B]Quinolin-12-Yl)Amino]-N-(4-Hydroxy-3-Methoxybenzyl)-6-Decenamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8U5, NAG, CL, MES, PEG |
 |
Crystal Structure Of Torpedo Californica Acetylcholinesterase In Complex With 2-{1-[4-(12-Amino-3-Chloro-6,7,10,11-Tetrahydro-7,11-Methanocycloocta[B]Quinolin-9-Yl)Butyl]-1H-1,2,3-Triazol-4-Yl}-N-[4-Hydroxy-3-Methoxybenzyl]Acetamide
Organism: Tetronarce californica
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8U2, PEG, CL |
 |
Crystal Structure Of Human Butyrylcholinesterase In Complex With 2-{1-[4-(12-Amino-3-Chloro-6,7,10,11-Tetrahydro-7,11-Methanocycloocta[B]Quinolin-9-Yl)Butyl]-1H-1,2,3-Triazol-4-Yl}-N-[4-Hydroxy-3-Methoxybenzyl]Acetamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2021-10-06 Classification: HYDROLASE Ligands: 8U2 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-10-06 Classification: VIRAL PROTEIN Ligands: SO4, CL, GOL |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-10-06 Classification: VIRAL PROTEIN Ligands: GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-06 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-10-06 Classification: VIRAL PROTEIN Ligands: GOL, PG4 |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-10-06 Classification: VIRAL PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-10-06 Classification: VIRAL PROTEIN Ligands: MPD, CIT, GOL |

