Search Count: 28
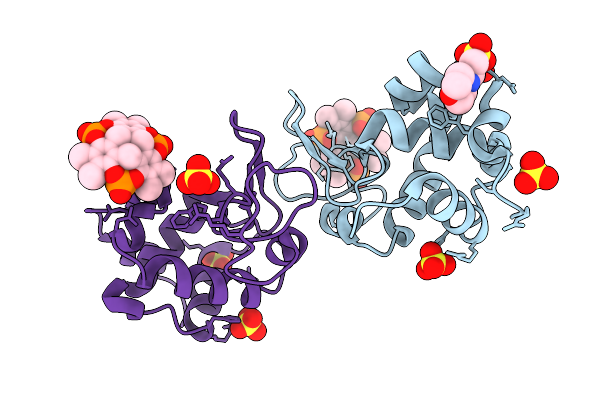 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: A1IXG, MES, SO4 |
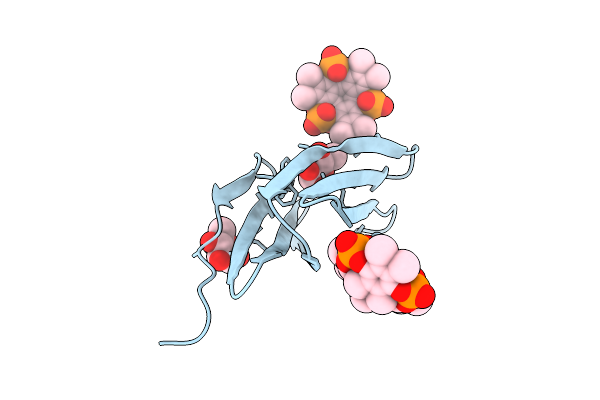 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IXG |
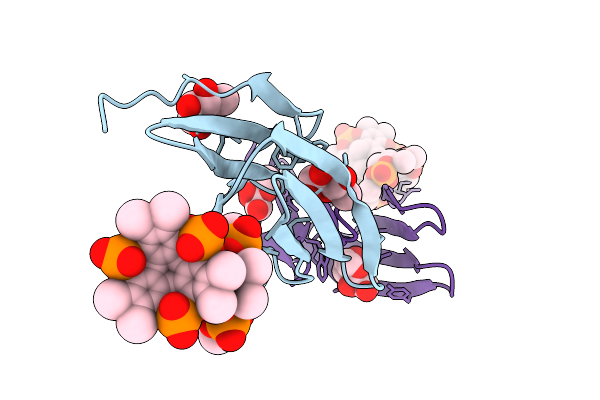 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IXG, CL |
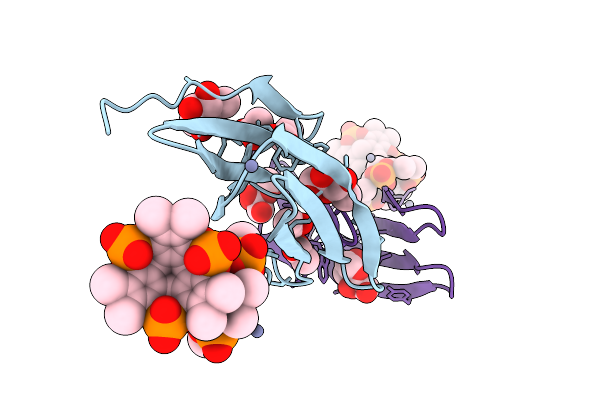 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: A1IXG, ACT, BDF, ZN |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IXG, ZN |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IXG |
 |
Organism: Thomasclavelia ramosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: PEG, ZN |
 |
Crystal Structure Of Bf3526 Peptidase From Bacteroides Fragilis In Complex With A Peptide
Organism: Bacteroides fragilis nctc 9343, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: K, ZN, EDO, POL, PEG, BU1, HEZ |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: EVB, BDF |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: T3Y, BDF |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: BDF, T3Y |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2025-03-05 Classification: SUGAR BINDING PROTEIN Ligands: BDF, T3Y |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-12-18 Classification: HYDROLASE Ligands: GTQ, EDO |
 |
Organism: Enterobacteria phage l1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-12-04 Classification: SUGAR BINDING PROTEIN Ligands: NDG, FWQ |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: BDF, EVB, GOL |
 |
The Rsl-D32N - Sulfonato-Calix[8]Arene Complex, I23 Form, Citrate Ph 4.0, Obtained By Cross-Seeding
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: EVB, BDF, GOL |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: GOL, BDF, EVB |
 |
Organism: Enterobacteria phage l1
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: EVB, NDG |
 |
Organism: Enterobacteria phage l1
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: NDG, EVB |
 |
Organism: Enterobacteria phage l1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-03-06 Classification: SUGAR BINDING PROTEIN Ligands: NDG |

