Search Count: 13
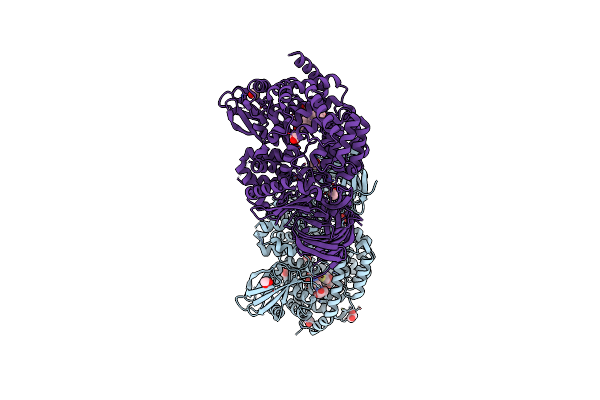 |
Erap1 In Complex With 1-[2-(3-Oxo-3,4-Dihydro-2H-1,4-Benzothiazin-4-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, B3P, EDO, BR, A1IMK |
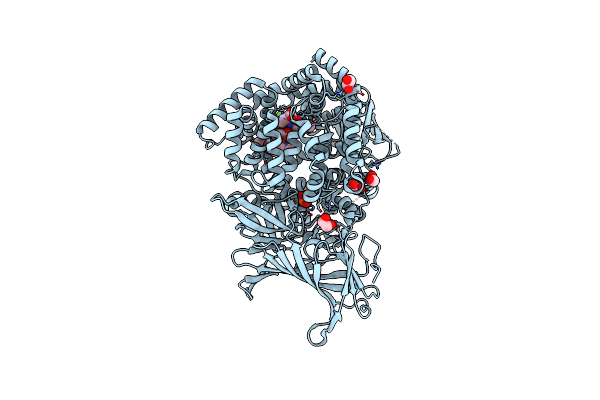 |
Erap1 In Complex With 1-[2-(6-Bromo-3-Oxo-3,4-Dihydro-2H-1,4-Benzoxazin-4-Yl)Acetamido]-4,4-Difluorocyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, PO4, EDO, A1IMJ |
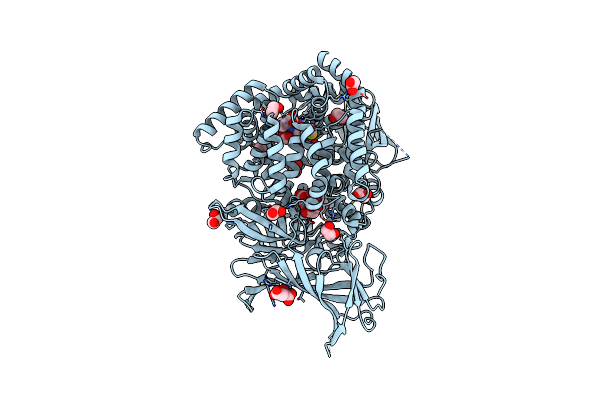 |
Erap1 In Complex With 1-[2-(6-Chloro-3-Oxo-3,4-Dihydro-2H-1,4-Benzothiazin-4-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, MLT, EDO, A1IMM |
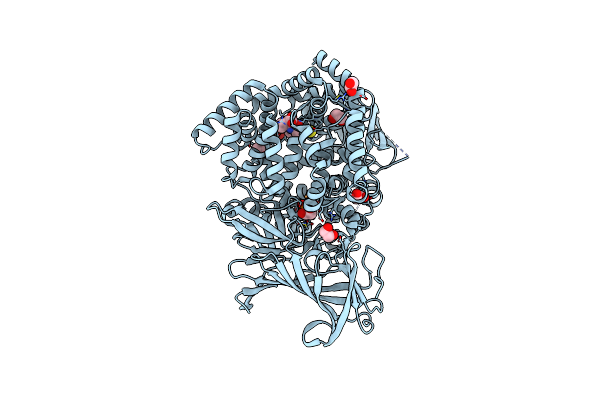 |
Erap1 In Complex With 1-[2-(2-Oxo-5-Phenyl-2,3-Dihydro-1,3-Benzothiazol-3-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, PO4, EDO, A1IML |
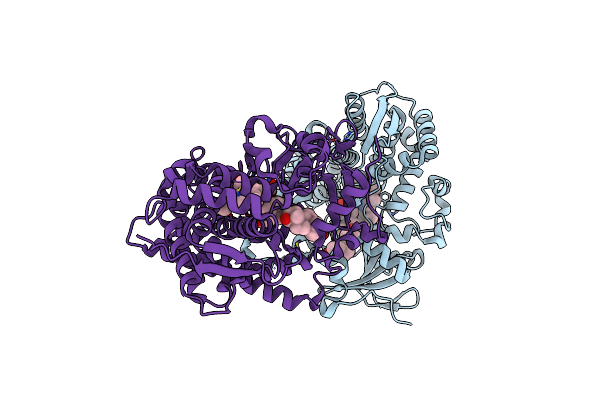 |
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2018-08-01 Classification: OXIDOREDUCTASE Ligands: HEM, ZN, TES, CL |
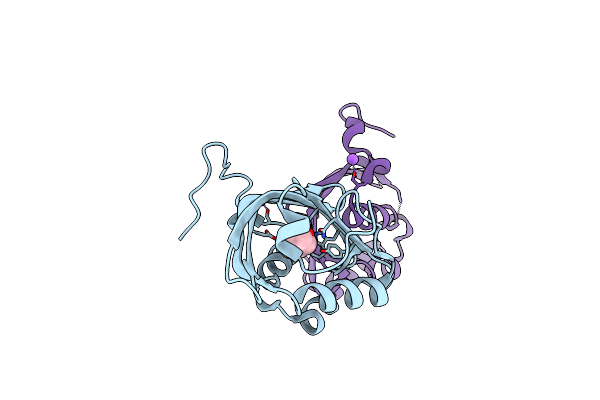 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2018-06-27 Classification: HYDROLASE Ligands: 3ZS, NA |
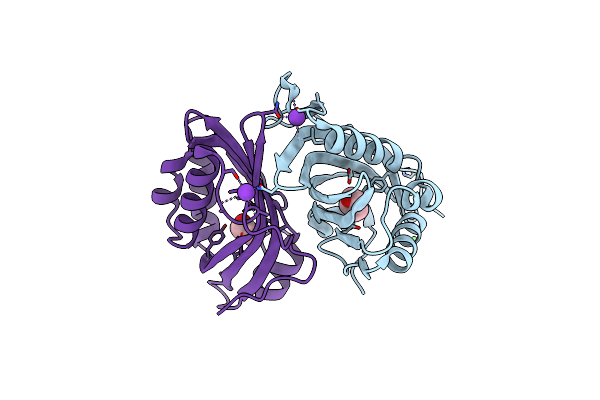 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-06-27 Classification: HYDROLASE Ligands: K, NI, 3ZS |
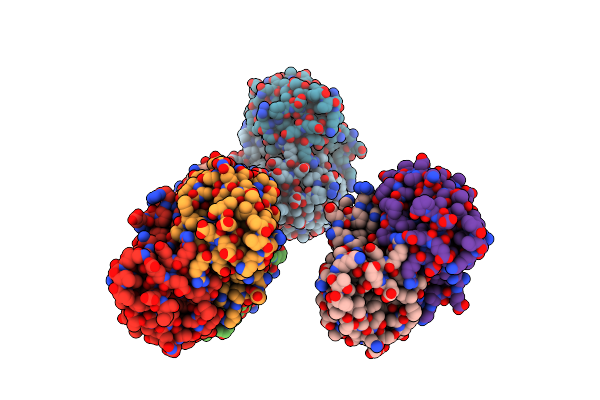 |
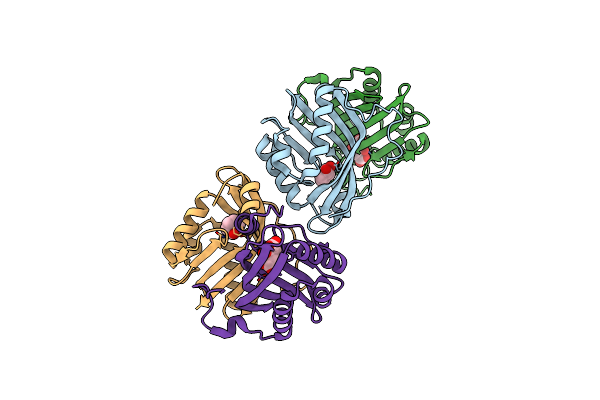
Crystal Structure Of The L74F/M78V/I80V/L114F Mutant Of Leh Complexed With Cyclopentene Oxide
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-06-27 Classification: HYDROLASE Ligands: 3ZS |
 |
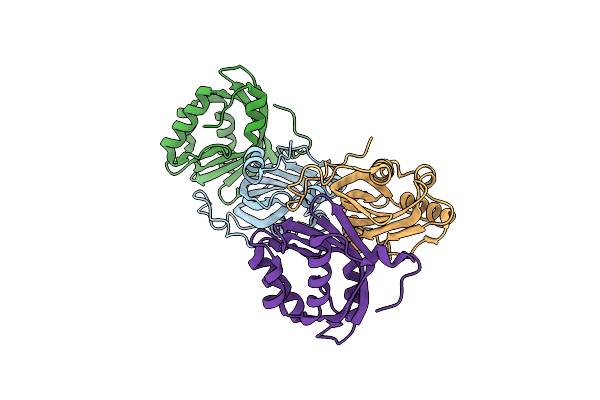
Crystal Structure Of The L74F/M78F/L103V/L114V/I116V/F139V/L147V Mutant Of Leh Complexed With (S,S)-Cyclohexanediol
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: 3ZQ, FLC |
 |
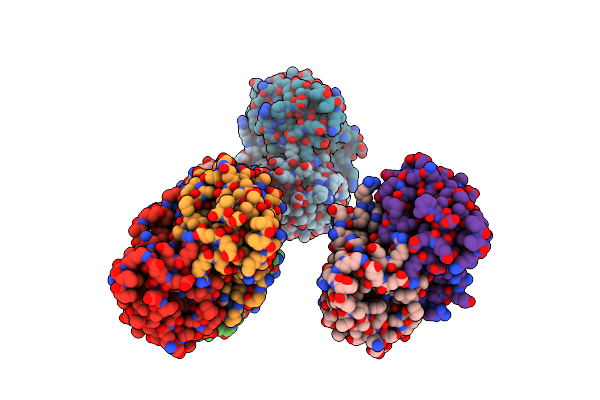
Crystal Structure Of The L74F/M78F/L103V/L114V/I116V/F139V/L147V Mutant Of Leh
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2015-07-15 Classification: HYDROLASE |
 |
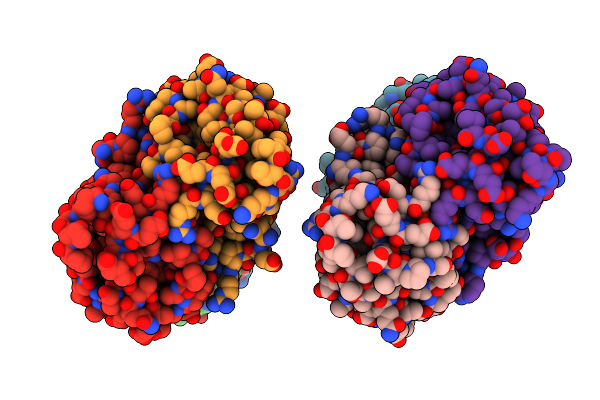
Crystal Structure Of The L74F/M78V/I80V/L114F Mutant Of Leh Complexed With Cyclopentene Oxide
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: 3ZS |
 |
Crystal Structure Of The L74F/M78V/I80V/L114F Mutant Of Leh Complexed With Cyclohexanediol
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: 40O |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-07-15 Classification: HYDROLASE |

