Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: CLR, Y01 |
 |
Cryo-Em Structure Of Nanodisc-Reconstituted Wildtype Human Mrp4 (In Complex With Vincristine)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: R1Q, ANP, MG |
 |
Cryo-Em Structure Of Nanodisc-Reconstituted Human Mrp4 Withe1202Q Mutation (In Complex With 5-Fluorouracil)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: URF, MG, ATP |
 |
Cryo-Em Structure Of Nanodisc-Reconstituted Human Mrp4 Withe1202Q Mutation (In Complex With Lapatinib)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: FMM, MG, ATP |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-03-05 Classification: TRANSCRIPTION |
 |
A 2.58A Crystal Structure Of S. Aureus Dna Gyrase And Dna With Metals Identified Through Anomalous Scattering
Organism: Staphylococcus aureus, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-10-23 Classification: ISOMERASE Ligands: GOL, MN, BTB |
 |
Cryo-Em Structure Of The Clpp Degradation System In Streptomyces Hawaiiensis
Organism: Streptomyces hawaiiensis
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: HYDROLASE |
 |
Cryo-Em Structure Of The Clpc1:Clpp1P2 Degradation Complex In Streptomyces Hawaiiensis
Organism: Streptomyces hawaiiensis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of The Clpc1:Clpp1P2 Degradation Complex In Streptomyces Hawaiiensis
Organism: Streptomyces hawaiiensis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Clpp1P2 In Complex With Adep1 From Streptomyces Hawaiiensis
Organism: Streptomyces hawaiiensis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: HYDROLASE |
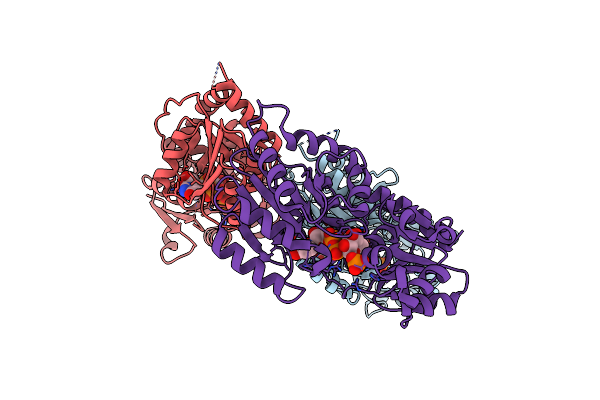 |
Structure Of Senb In Complex With Udp-Glc And Po4- At 1.98 Angstroms Resolution
Organism: Variovorax paradoxus
Method: X-RAY DIFFRACTION Release Date: 2024-02-07 Classification: TRANSFERASE Ligands: UPG, PO4 |
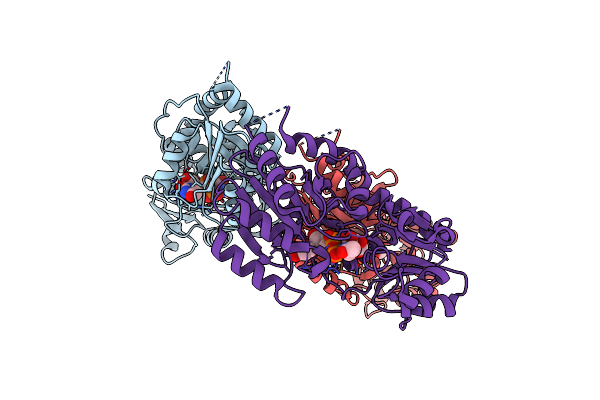 |
Organism: Variovorax paradoxus
Method: X-RAY DIFFRACTION Release Date: 2024-02-07 Classification: TRANSFERASE Ligands: UD2 |
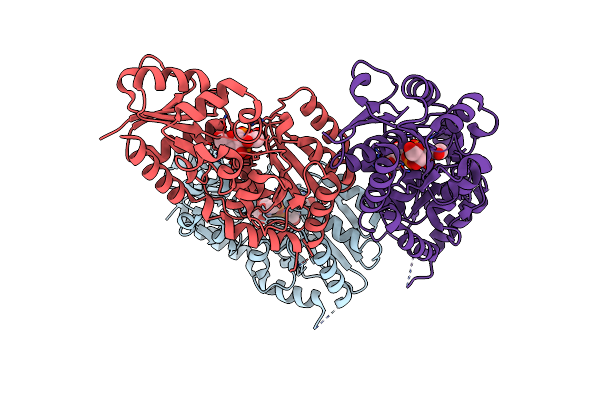 |
Organism: Variovorax paradoxus
Method: X-RAY DIFFRACTION Release Date: 2024-02-07 Classification: TRANSFERASE Ligands: UD1 |
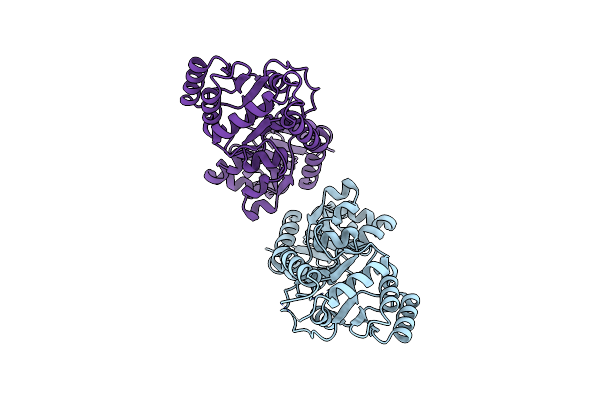 |
Organism: Ramlibacter sp.
Method: X-RAY DIFFRACTION Release Date: 2024-02-07 Classification: TRANSFERASE |
 |
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: UDP, GOL |
 |
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: 6JI, UDP, TRS |
 |
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: Q30, UDP, TRS |
 |
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: UDP, TRS, BUF |
 |
Organism: Geobacter sulfurreducens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: ANTIVIRAL PROTEIN Ligands: NAD |
 |
Organism: Geobacter sulfurreducens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: ANTIVIRAL PROTEIN/DNA/RNA Ligands: NAD |