Search Count: 16
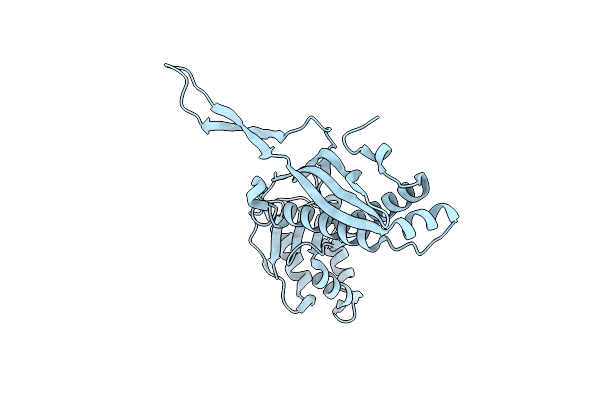 |
Organism: Arthrobacter sp.
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-05-21 Classification: CYTOSOLIC PROTEIN |
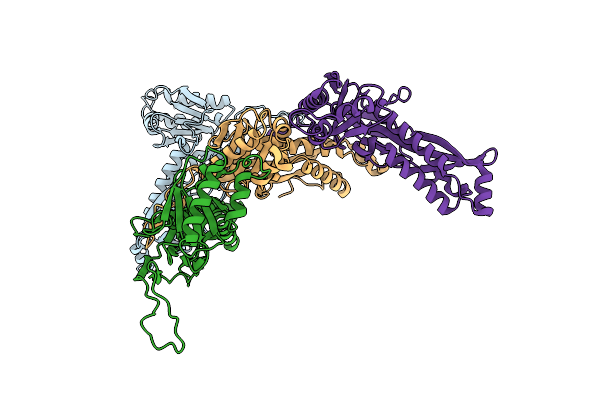 |
Organism: Dendrosporobacter quercicolus
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-05-21 Classification: CYTOSOLIC PROTEIN |
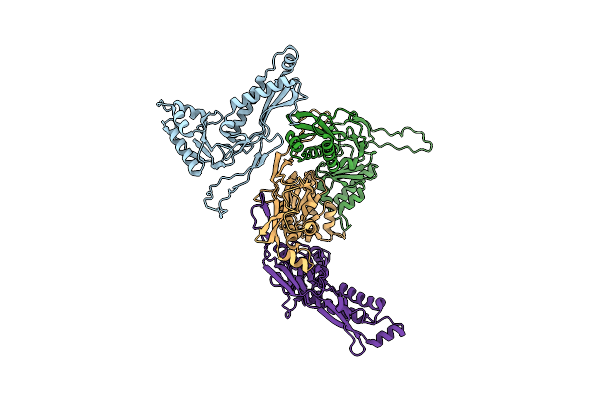 |
Organism: Dendrosporobacter quercicolus
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2025-05-21 Classification: CYTOSOLIC PROTEIN |
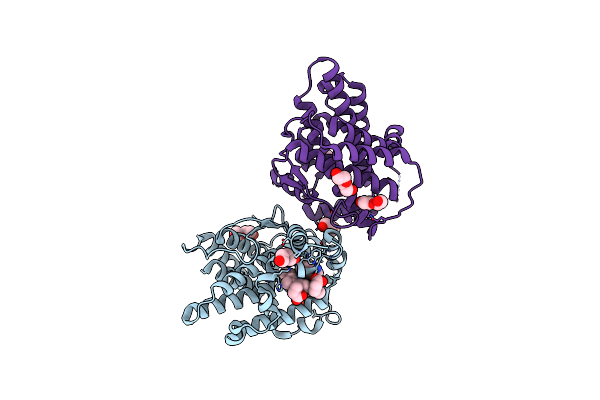 |
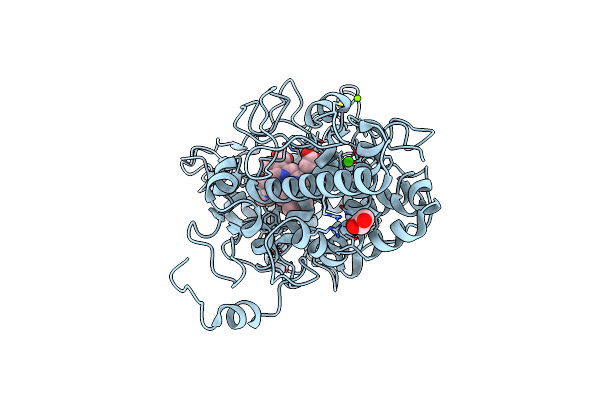
Heme-Dependent L-Tyrosine Hydroxylase (Tyrh) From Streptomyces Sclerotialus: Fourfold Mutant
Organism: Streptomyces sclerotialus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: HEM, IND, PEG, ACT |
 |
Organism: Cyanobacterium tdx16
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, CA, MG |
 |
Organism: Thermus caliditerrae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: MG |
 |
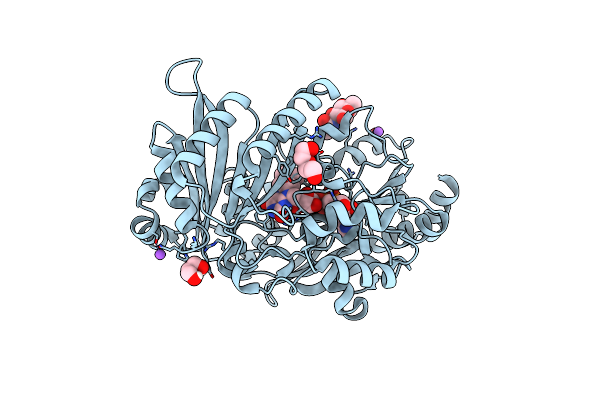
Short-Chain Dehydrogenase/Reductase (Sdr) From Thermus Caliditerrae In Complex With Nadp
Organism: Thermus caliditerrae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: NAP, MG |
 |
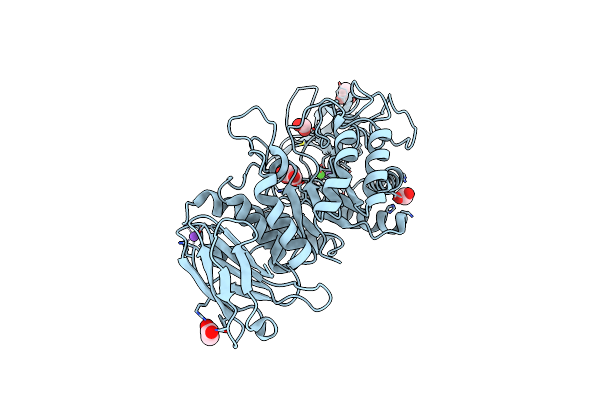
Organism: Citrus sinensis
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-09-06 Classification: FLAVOPROTEIN Ligands: FAD, NAG, CL, IOD, NA, GOL, PEG |
 |
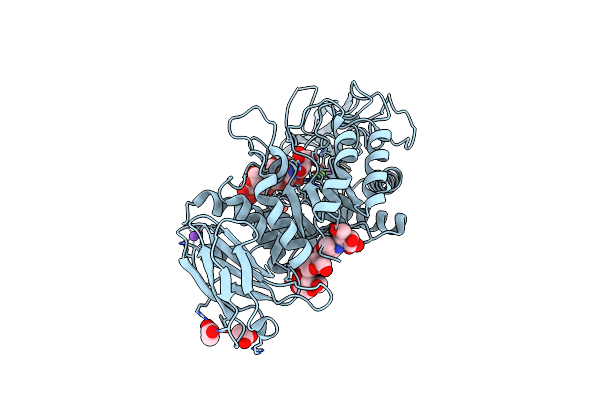
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CA, NA, MLI, FMT, ACY |
 |
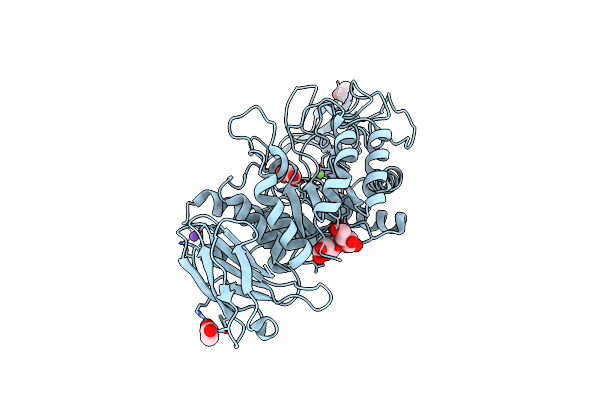
Crystal Structure Of Bacillus Paralicheniformis Alpha-Amylase In Complex With Acarbose
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CA, NA, ACY |
 |
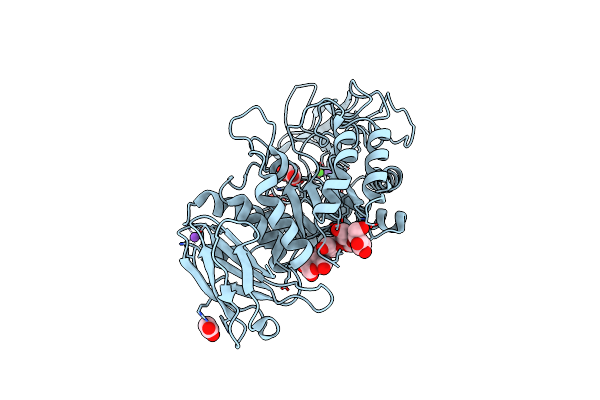
Crystal Structure Of Bacillus Paralicheniformis Alpha-Amylase In Complex With Maltose
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CA, NA, MLI, ACY |
 |
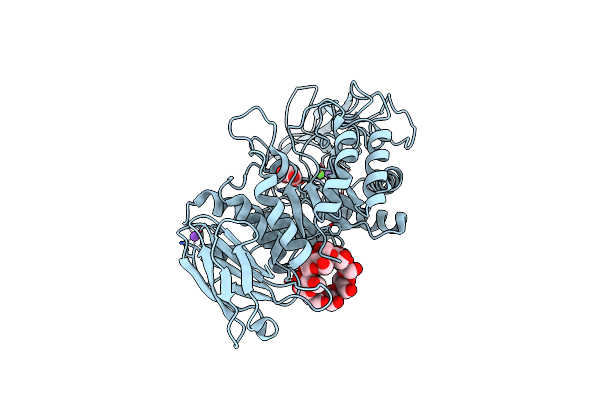
Crystal Structure Of Bacillus Paralicheniformis Alpha-Amylase In Complex With Maltotetraose
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CA, NA, MLI, ACY |
 |
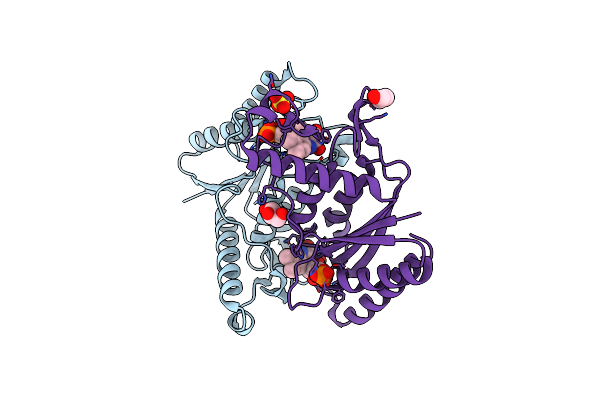
Crystal Structure Of Bacillus Paralicheniformis Alpha-Amylase In Complex With Beta-Cyclodextrin
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-10-14 Classification: HYDROLASE Ligands: CA, NA, MLI |
 |
Organism: Bacillus okhensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, SO4 |
 |
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-12-04 Classification: VIRUS LIKE PARTICLE Ligands: SO4, GOL |
 |
Structure Of Nitrincola Lacisaponensis Flavin-Containing Monooxygenase (Fmo) In Complex With Fad And Nadp+
Organism: Nitrincola lacisaponensis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-01-02 Classification: FLAVOPROTEIN Ligands: FAD, NAP, MG |

