Search Count: 17
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: SIGNALING PROTEIN Ligands: MG, AF3, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: SIGNALING PROTEIN Ligands: ZN, MG, AF3, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture -Pip2/+Sterol (Dopc, Dope, Dops, Cholesterol 30:20:20:30)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture +Pip2/-Sterol (Dopc, Dope, Dops, Pi(4,5)P2 50:20:20:10)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN Ligands: I3P |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture +Pip2/+Sterol (Dopc, Dope, Dops, Cholesterol, Pi(4,5)P2 35:20:20:15:10)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN Ligands: I3P, P5S |
 |
Compact State - Pil1 In Native Eisosome Lattice Bound To Plasma Membrane Microdomain
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Compact State - Pil1 Dimer With Lipid Headgroups Fitted In Native Eisosome Lattice Bound To Plasma Membrane Microdomain
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN Ligands: I3P, SEP |
 |
Stretched State - Pil1 In Native Eisosome Lattice Bound To Plasma Membrane Microdomain
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Toroid (Torc1 Organized In Inhibited Domains).
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: SIGNALING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: SIGNALING PROTEIN |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Target Of Rapamycin Complex 2
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Resolution:7.90 Å Release Date: 2017-12-06 Classification: SIGNALING PROTEIN |
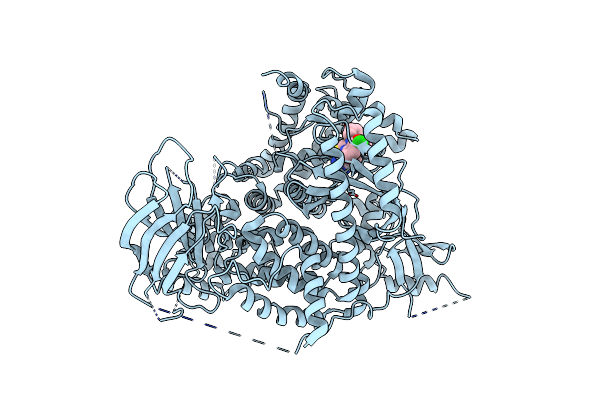 |
A Pharmacological Map Of The Pi3-K Family Defines A Role For P110 Alpha In Signaling: The Structure Of Complex Of Phosphoinositide 3- Kinase Gamma With Inhibitor Pik-39
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-05-22 Classification: TRANSFERASE/INHIBITOR Ligands: 039 |
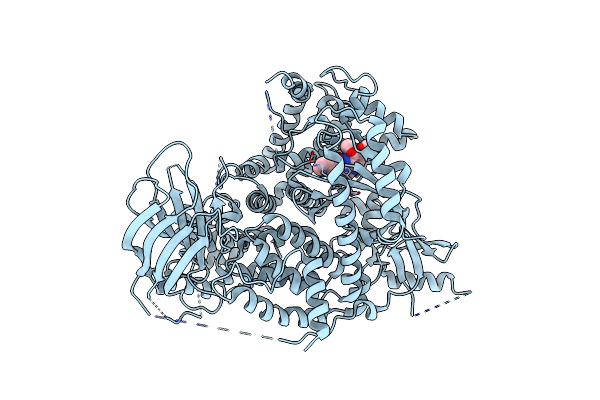 |
A Pharmacological Map Of The Pi3-K Family Defines A Role For P110Alpha In Signaling: The Structure Of Complex Of Phosphoinositide 3-Kinase Gamma With Inhibitor Pik-90
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-05-22 Classification: TRANSFERASE Ligands: 090 |
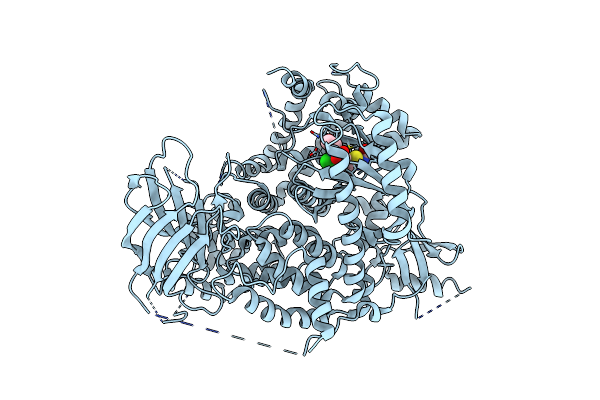 |
A Pharmacological Map Of The Pi3-K Family Defines A Role For P110Alpha In Signaling: The Structure Of Complex Of Phosphoinositide 3-Kinase Gamma With Inhibitor Pik-93
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-05-22 Classification: TRANSFERASE Ligands: 093 |

