Search Count: 16
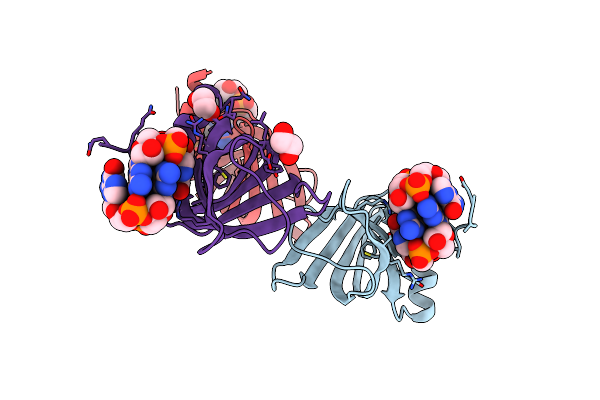 |
Structure Of The Alg44 Pilz Domain From Pseudomonas Aeruginosa Pao1 In Complex With C-Di-Gmp
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-04-08 Classification: PROTEIN BINDING Ligands: C2E, EDO |
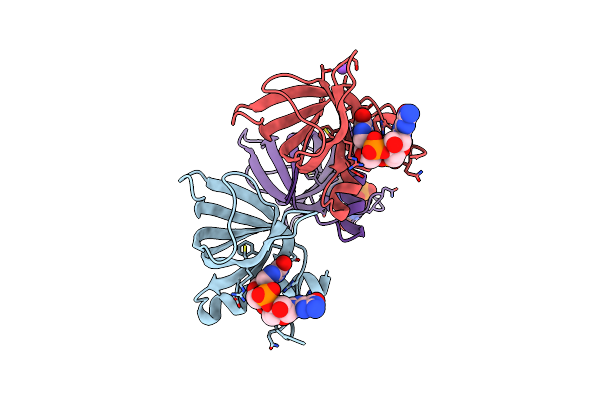 |
Structure Of The Alg44 Pilz Domain (R95A Mutant) From Pseudomonas Aeruginosa Pao1 In Complex With C-Di-Gmp
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-04-08 Classification: PROTEIN BINDING Ligands: C2E, CL, NA |
 |
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2011-12-21 Classification: DE NOVO PROTEIN Ligands: BTB, GOL, NA |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2010-07-21 Classification: Carbohydrate-binding protein |
 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2008-03-25 Classification: HYDROLASE Ligands: CA, GOL |
 |
Penicillium Citrinum Alpha-1,2-Mannosidase In Complex With A Substrate Analog
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-03-25 Classification: HYDROLASE Ligands: CA, GOL |
 |
Organism: Anas platyrhynchos
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-09-07 Classification: LYASE |
 |
Organism: Anas platyrhynchos
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-09-07 Classification: LYASE |
 |
Crystal Structure Of T161D Duck Delta 2 Crystallin Mutant With Bound Argininosuccinate
Organism: Anas platyrhynchos
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-09-07 Classification: LYASE Ligands: AS1 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2004-05-04 Classification: TRANSFERASE Ligands: CL, GOL |
 |
Crystal Structure Of Yeast Alpha1,2-Mannosyltransferase Kre2P/Mnt1P: Binary Complex With Gdp/Mn
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2004-05-04 Classification: TRANSFERASE Ligands: MN, CL, GDP, EPE |
 |
Crystal Structure Of Yeast Alpha1,2-Mannosyltransferase Kre2P/Mnt1P: Ternary Complex With Gdp/Mn And Methyl-Alpha-Mannoside Acceptor
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2004-05-04 Classification: TRANSFERASE Ligands: MMA, MN, CL, GDP, EPE |
 |
Structure Of P. Citrinum Alpha 1,2-Mannosidase Reveals The Basis For Differences In Specificity Of The Er And Golgi Class I Enzymes
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-02-20 Classification: HYDROLASE Ligands: CA, DMJ |
 |
Structure Of P. Citrinum Alpha 1,2-Mannosidase Reveals The Basis For Differences In Specificity Of The Er And Golgi Class I Enzymes
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-02-20 Classification: HYDROLASE Ligands: CA, KIF |
 |
Structure Of P. Citrinum Alpha 1,2-Mannosidase Reveals The Basis For Differences In Specificity Of The Er And Golgi Class I Enzymes
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-01-23 Classification: HYDROLASE Ligands: CA |
 |
X-Ray Crystal Structure Of The Human Dimeric S-Lac Lectin, L-14-Ii, In Complex With Lactose At 2.9 Angstroms Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1994-04-30 Classification: LECTIN |

