Search Count: 16
 |
Crystal Structure Of The Human Cu, Zn Superoxide Dismutase, Familial Amyotrophic Lateral Sclerosis (Fals) Mutant H43R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-09-09 Classification: OXIDOREDUCTASE Ligands: CU1, ZN, SO4 |
 |
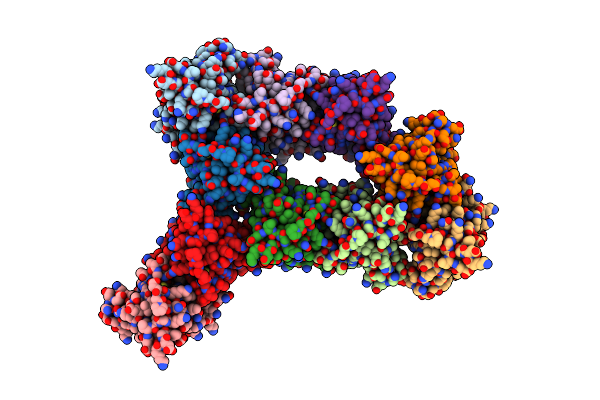
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-09-09 Classification: OXIDOREDUCTASE Ligands: CU1, ZN, SO4 |
 |
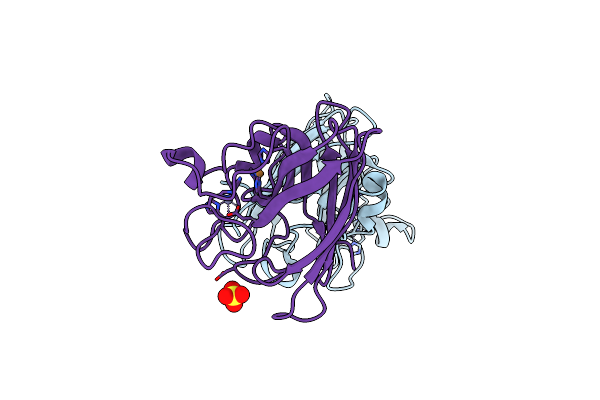
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-11-27 Classification: OXIDOREDUCTASE Ligands: CU1, ZN, SO4 |
 |
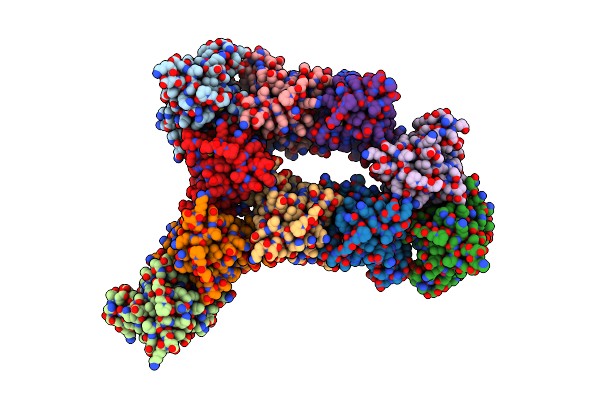
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2002-11-27 Classification: OXIDOREDUCTASE Ligands: CU1, ZN, SO4 |
 |
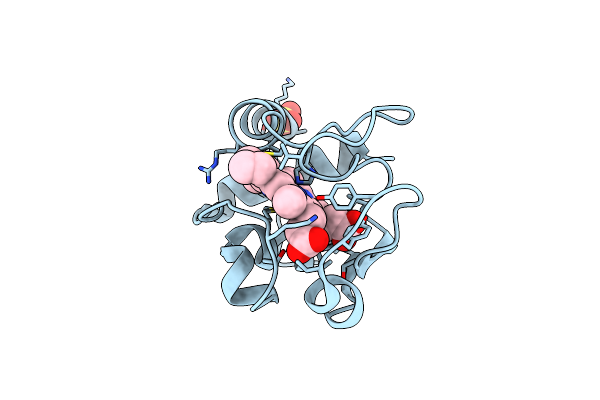
Superoxide Dismutase Mutant With Lys 136 Replaced By Glu, Cys 6 Replaced By Ala And Cys 111 Replaced By Ser (K136E, C6A, C111S)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 1999-07-23 Classification: OXIDOREDUCTASE Ligands: CU, ZN, SO4 |
 |
Structural And Functional Effects Of Multiple Mutations At Distal Sites In Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-01-26 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Structural And Functional Effects Of Multiple Mutations At Distal Sites In Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-01-26 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Structural And Functional Effects Of Multiple Mutations At Distal Sites In Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-01-26 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Structural And Functional Effects Of Multiple Mutations At Distal Sites In Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-01-26 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Replacements In A Conserved Leucine Cluster In The Hydrophobic Heme Pocket Of Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 1995-01-26 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Replacements In A Conserved Leucine Cluster In The Hydrophobic Heme Pocket Of Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-01-26 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Replacements In A Conserved Leucine Cluster In The Hydrophobic Heme Pocket Of Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-01-26 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Replacements In A Conserved Leucine Cluster In The Hydrophobic Heme Pocket Of Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1995-01-26 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Structural Studies Of The Roles Of Residues 82 And 85 At The Interactive Face Of Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 1994-12-20 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Structural Studies Of The Roles Of Residues 82 And 85 At The Interactive Face Of Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1994-12-20 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |
 |
Structural Studies Of The Roles Of Residues 82 And 85 At The Interactive Face Of Cytochrome C
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1994-12-20 Classification: ELECTRON TRANSPORT(HEME PROTEIN) Ligands: SO4, HEC |

