Search Count: 176
 |
Crystal Structure Of Kirsten Rat Sarcoma G12C Complexed With Gmppnp And Covalently Bound To An Adduct Of {(2S)-4-[7-(8-Chloronaphthalen-1-Yl)-2-{[(2S)-1-Methylpyrrolidin-2-Yl]Methoxy}-5,6,7,8-Tetrahydropyrido[3,4-D]Pyrimidin-4-Yl]-1-[(2Z)-2-Fluoro-3-(Pyridin-2-Yl)Prop-2-Enoyl]Piperazin-2-Yl}Acetonitrile
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GNP, A1B7P, MG |
 |
Crystal Structure Of Kirsten Rat Sarcoma G12C Complexed With Gdp And Covalently Bound To An Adduct Of (2S)-1-{4-[(7P)-7-(8-Ethynyl-7-Fluoro-3-Hydroxynaphthalen-1-Yl)-8-Fluoro-2-{[(2R,4R,7As)-2-Fluorotetrahydro-1H-Pyrrolizin-7A(5H)-Yl]Methoxy}Pyrido[4,3-D]Pyrimidin-4-Yl]Piperazin-1-Yl}-2-Fluoro-3-(1,3-Thiazol-2-Yl)Propan-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, A1B7Q, MG |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN |
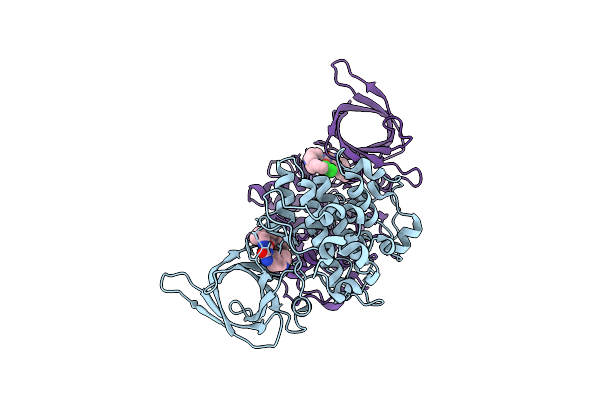 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: A1IXL |
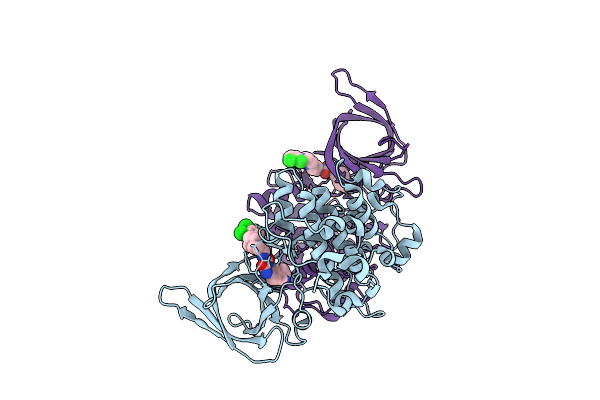 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: A1IXM |
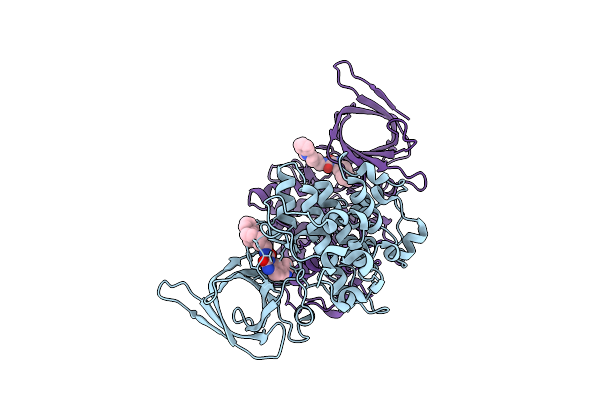 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: A1IXN |
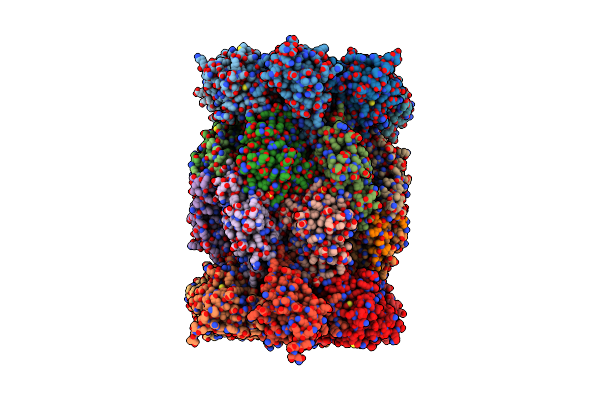 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: SO4, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: SO4, MG, A1AU6 |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-04-30 Classification: TRANSPORT PROTEIN Ligands: CA, GOL, A1ILE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.42 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: MG, SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: SO4, MG, A1A9B |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: SO4, A1A9B, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: SO4, MG, A1AHA, MPD, ACY |
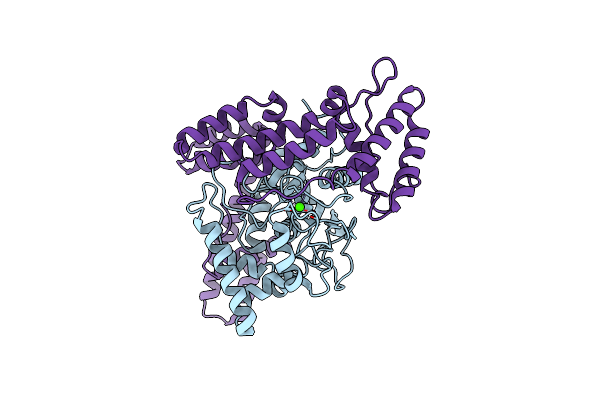 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.17 Å Release Date: 2025-02-19 Classification: LIPID BINDING PROTEIN Ligands: CA, CL |
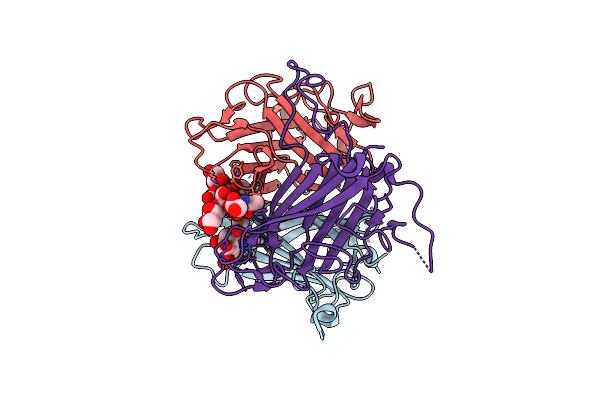 |
Human Adenovirus Type 36 Fiber Knob In Complex With 4-O-Acetyl-3'-Sialyllactose
Organism: Human adenovirus 36
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: PKM |
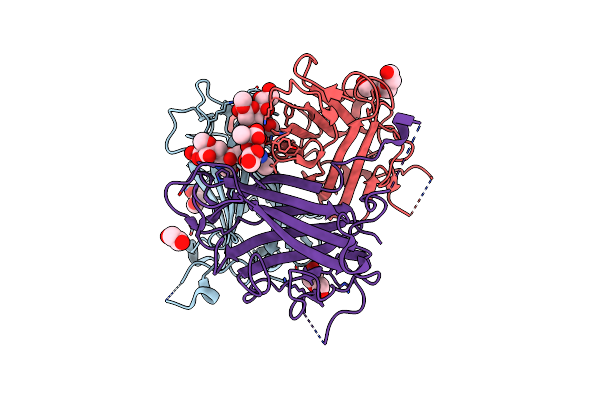 |
Human Adenovirus Type 36 Fiber Knob In Complex With 4-O,5-N-Diacetylneuraminic Acid
Organism: Human adenovirus 36
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: ANA, PEG, PGE, EDO |
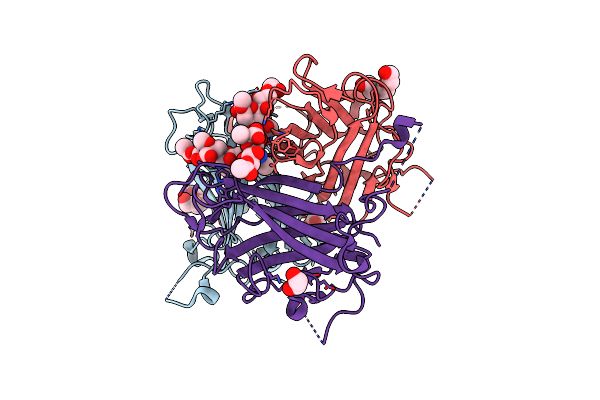 |
Human Adenovirus Type 36 Fiber Knob In Complex With 4,9-O,5-N-Triacetylneuraminic Acid
Organism: Human adenovirus 36
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: SIO, PGE, EDO |
 |
Human Adenovirus Type 37 Fiber Knob In Complex With 4-O,5-N-Diacetylneuraminic Acid
Organism: Human adenovirus d37
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: ANA, ZN, ACT, CL |
 |
Crystal Structure Of A Fatty Acid Decarboxylase From Kocuria Marina In Complex With Myristic Acid
Organism: Kocuria marina
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: MYR, HEM, GOL, PEG |

