Search Count: 214
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN Ligands: A2G |
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN Ligands: GOL, A2G |
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Staphylococcus Aureus Cysteine-Free Scda With Bound Iron, Determined By Molecular Replacement And Fe Anomalous Signal
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: METAL BINDING PROTEIN Ligands: FE, O, ZN |
 |
Cryo-Em Structure Of Anp Amyloids From Left Atrial Appendage Of Atrial Fibrillation Patient - Polymorph A
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: PROTEIN FIBRIL |
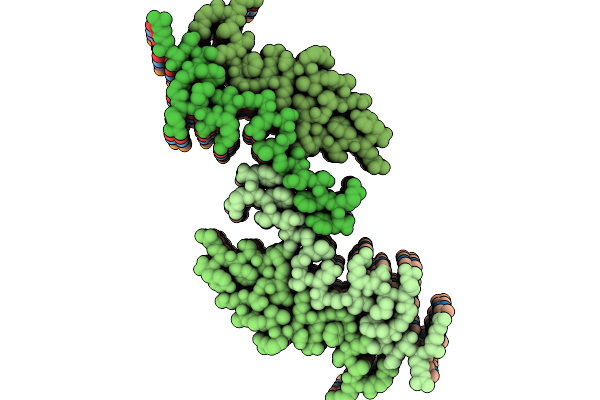 |
Cryo-Em Structure Of Anp Amyloids From Left Atrial Appendage Of Atrial Fibrillation Patient - Polymorph B
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: PROTEIN FIBRIL |
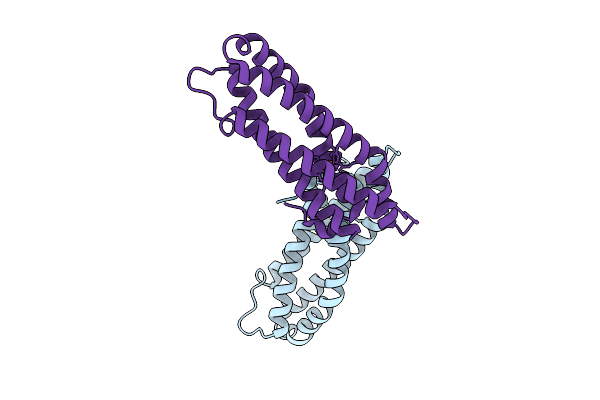 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXYGEN BINDING Ligands: FE, O |
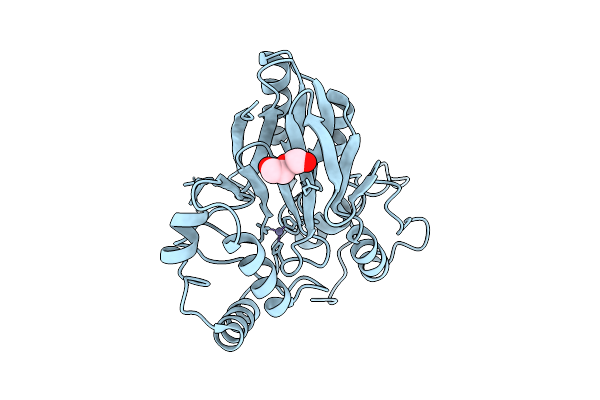 |
Crystal Structure Of The Persulfide Dioxygenase (Pdo - Pa2915) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: PEG, ZN, CL |
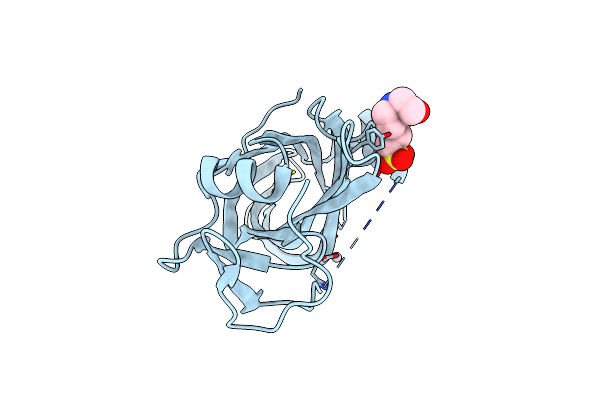 |
Crystal Structure Of Protein D: Defoliating Toxin Form Fusarium Oxysporum F.Sp. Vasinfectum
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-05-28 Classification: TOXIN Ligands: EPE, HG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-02-12 Classification: SUGAR BINDING PROTEIN Ligands: SER, GOL, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2025-02-12 Classification: SUGAR BINDING PROTEIN Ligands: THR |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of Rnase A With [Ru2(D-P-Fphf)(O2Cch3)2(O2Co)] Complex
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN Ligands: SO4, A1IQW |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of Rnase A With [Ru2Cl(D-P-Cnphf)(O2Cch3)3]
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: SO4, A1IHW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The Gluk1 Ligand-Binding Domain In Complex With Kainate And Bpam538 At 1.90 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: KAI, SO4, CL, GOL, 9TE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: RECOMBINATION |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cc25.56
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-08-07 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cc25.4
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: IMMUNE SYSTEM Ligands: MPD, NAG |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cc25.43
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: IMMUNE SYSTEM Ligands: NAG |

