Search Count: 54
 |
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN |
 |
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN |
 |
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Transcription Factor Ntca From Synechococcus Elongatus In Complex With Its Target Dna.
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN Ligands: AKG |
 |
Crystal Structure Of Transcription Factor Ntca From Synechococcus Elongatus In Complex With Its Transcriptional Co- Activator Pipx And Its Target Dna (Crystal Ii)
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:4.30 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN Ligands: AKG |
 |
Crystal Structure Of Transcription Factor Ntca From Synechococcus Elongatus In Complex With Its Transcriptional Co- Activator Pipx And Its Target Dna (Crystal I)
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2025-03-12 Classification: DNA BINDING PROTEIN Ligands: AKG |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Swivelled Conformation (Model Py48S-Auc-Swiv-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2.1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-Eif5)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, MET, GCP |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2023-09-27 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Sars-Cov-2 S Protein S:D614G Mutant In 3-Down With Binding Site Of An Entry Inhibitor
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Resolution:4.30 Å Release Date: 2023-09-27 Classification: VIRAL PROTEIN Ligands: NAG, NA, XIO |
 |
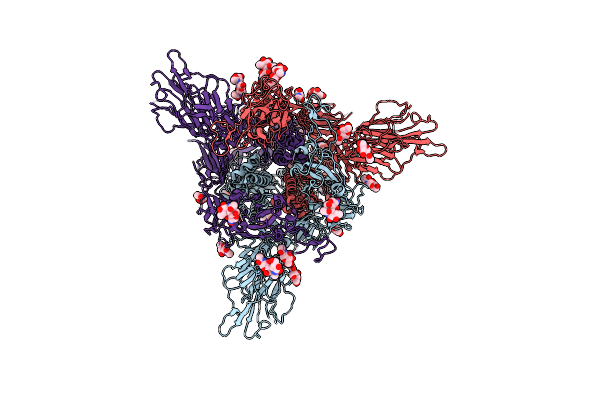
Organism: Severe acute respiratory syndrome coronavirus 2, Escherichia virus t4
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2022-05-25 Classification: VIRAL PROTEIN Ligands: NAG |
 |
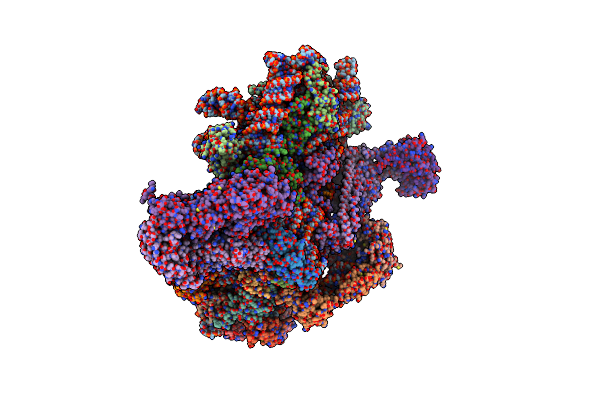
Organism: Severe acute respiratory syndrome coronavirus 2, Escherichia virus t4
Method: ELECTRON MICROSCOPY Resolution:4.20 Å Release Date: 2022-05-25 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure Of A Partial Yeast 48S Preinitiation Complex In Open Conformation.
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Resolution:5.15 Å Release Date: 2019-07-31 Classification: RIBOSOME Ligands: 7NO, MG, ZN, GCP |

