Search Count: 1,381
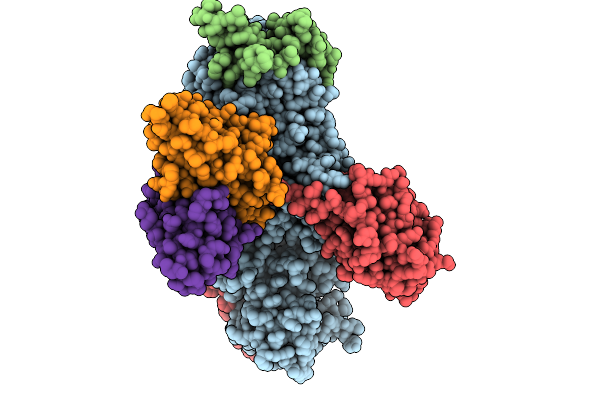 |
Organism: Human gammaherpesvirus 4, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
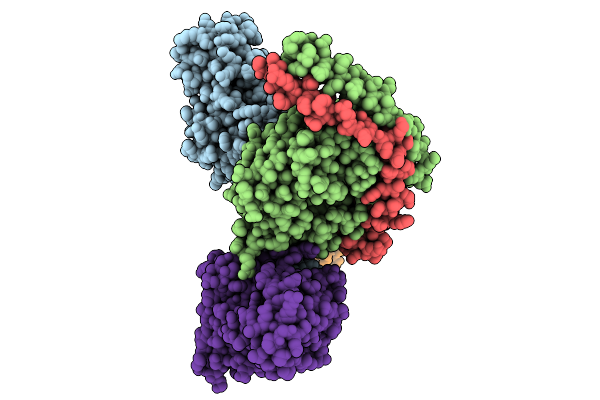 |
Consensus Olfactory Receptor Consor6 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:2.78 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EJQ, CLR |
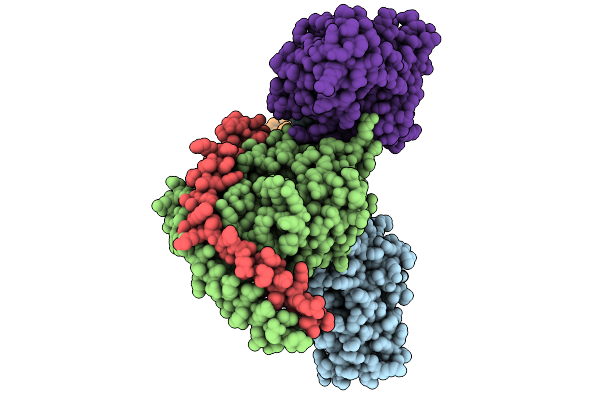 |
Consensus Olfactory Receptor Consor6 Bound To Alpha-Hexyl Cinnamaldehyde And In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:2.62 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: CLR, A1EJG |
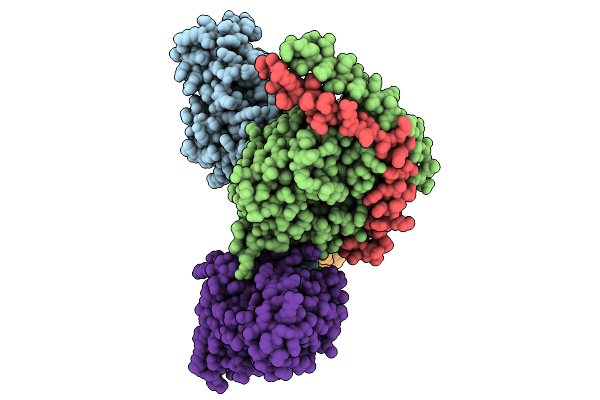 |
Consensus Olfactory Receptor Consor6 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:2.83 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: CLR, BZQ |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.17 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: A1EJQ, CLR |
 |
Consensus Olfactory Receptor Bmor6A2 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Mus musculoides, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.54 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: OYA |
 |
Consensus Olfactory Receptor Bmor6A2 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EJH |
 |
Consensus Olfactory Receptor Bmor6A2 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:3.33 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EJH |
 |
Organism: Homo sapiens, Niallia circulans
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2026-01-21 Classification: STRUCTURAL PROTEIN Ligands: A1L8O |
 |
Complex Of Fmdv Asia1/Js/05 And Porcine-Derived Neutralizing Monoclonal Antibody Pas12
Organism: Foot-and-mouth disease virus asia 1, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:2.14 Å Release Date: 2026-01-21 Classification: VIRUS/IMMUNE SYSTEM |
 |
Complex Of Fmdv Asia1/Js/05 And Porcine-Derived Neutralizing Monoclonal Antibody Pas5
Organism: Foot-and-mouth disease virus asia 1, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:2.17 Å Release Date: 2026-01-21 Classification: VIRUS/IMMUNE SYSTEM |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2026-01-21 Classification: TRANSPORT PROTEIN Ligands: IHP |
 |
Organism: Mus musculus, Murid herpesvirus 68
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Murid gammaherpesvirus 4
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN |
 |
Organism: Macacine gammaherpesvirus 4, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus, Semliki forest virus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS Ligands: CA |
 |
Organism: Semliki forest virus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Streptococcus macedonicus
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2026-01-07 Classification: UNKNOWN FUNCTION |
 |
Organism: African swine fever virus
Method: ELECTRON MICROSCOPY Resolution:2.92 Å Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN |

