Search Count: 29
 |
Crystal Structure Of A Cyclodipeptide Synthase From Streptomyces Sapporonensis
Organism: Streptomyces cinnamoneus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-04 Classification: LIGASE Ligands: EDO, GOL, PEG |
 |
Organism: Thermoanaerobacter tengcongensis (strain dsm 15242 / jcm 11007 / nbrc 100824 / mb4)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: PO4, MG |
 |
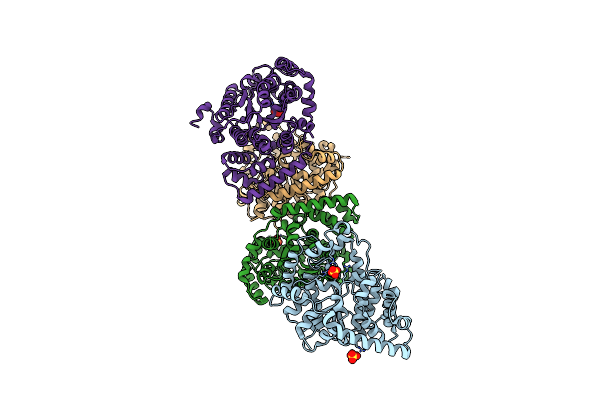
Non-Specific And Specific Interactions Work Cooperatively To Promote Cytidine Deamination Catalyzed By Apobec3A
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2021-10-06 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Streptococcus thermophilus lmg 18311
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2021-07-21 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of The Hiv Capsid Hexamer Bound To The Small Molecule Long-Acting Inhibitor, Gs-6207
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-01 Classification: VIRAL PROTEIN Ligands: QNG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: MN, AKG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: MN, EOH |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of The Recombination Mediator Protein Recf-Atprs In Recfor Pathway
Organism: Caldanaerobacter subterraneus subsp. tengcongensis (strain dsm 15242 / jcm 11007 / nbrc 100824 / mb4)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-10-17 Classification: DNA BINDING PROTEIN Ligands: AGS |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis (strain dsm 15242 / jcm 11007 / nbrc 100824 / mb4)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-04-18 Classification: DNA BINDING PROTEIN |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis (strain dsm 15242 / jcm 11007 / nbrc 100824 / mb4)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-04-18 Classification: DNA BINDING PROTEIN Ligands: ATP, IMD, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2017-12-20 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-12-20 Classification: TRANSCRIPTION |
 |
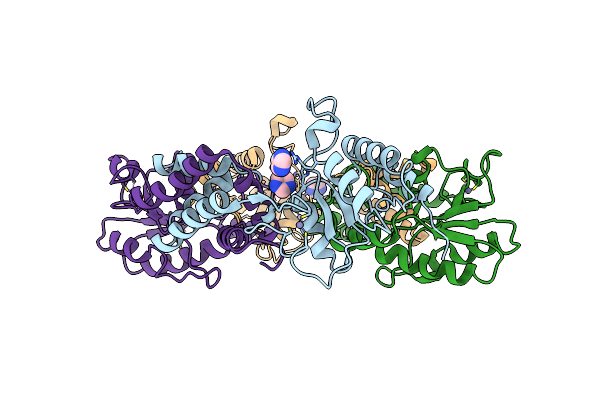
Structural Basis Underlying Complex Assembly Andconformational Transition Of The Type I R-M System
Organism: Caldanaerobacter subterraneus subsp. tengcongensis (strain dsm 15242 / jcm 11007 / nbrc 100824 / mb4), Caldanaerobacter subterraneus subsp. tengcongensis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2017-11-29 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
 |
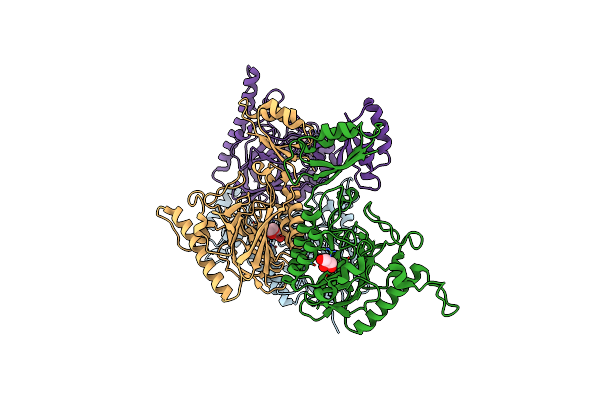
Structural And Functional Studies The Characterization Of Cys4 Zinc-Finger Motif In The Recombination Mediator Protein Recr
Organism: Thermoanaerobacter tengcongensis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-12-10 Classification: RECOMBINATION Ligands: ZN, IMD |
 |
Structural And Functional Studies The Characterization Of C58G/C70G Mutant In Cys4 Zinc-Finger Motif In The Recombination Mediator Protein Recr
Organism: Thermoanaerobacter tengcongensis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-12-10 Classification: RECOMBINATION Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-08-27 Classification: HYDROLASE Ligands: GOL |
 |
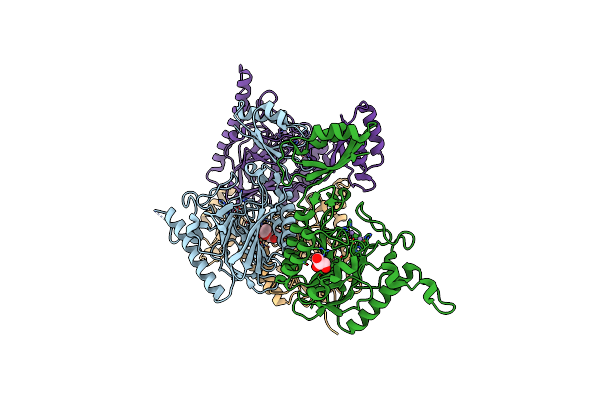
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-02-26 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2014-02-26 Classification: HYDROLASE Ligands: GOL, MN |
 |
Organism: Acidovorax citrulli
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2013-10-23 Classification: LYASE Ligands: CA |

