Search Count: 85
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-07-09 Classification: GENE REGULATION |
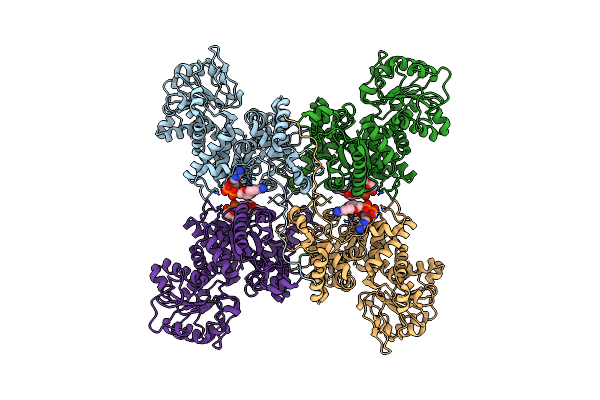 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In Apo Form
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD |
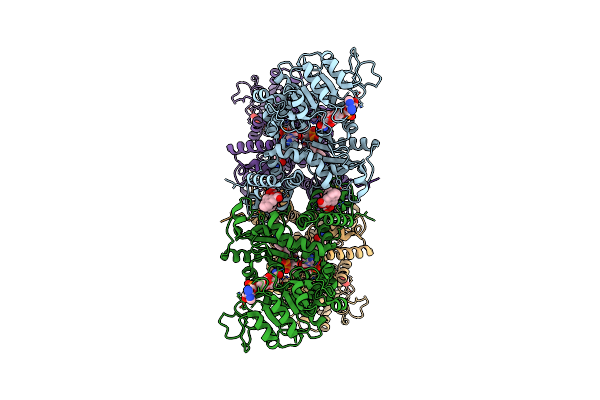 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In A Ternary Complex With Nad+ And Allosteric Inhibitor Ea
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD, JBR |
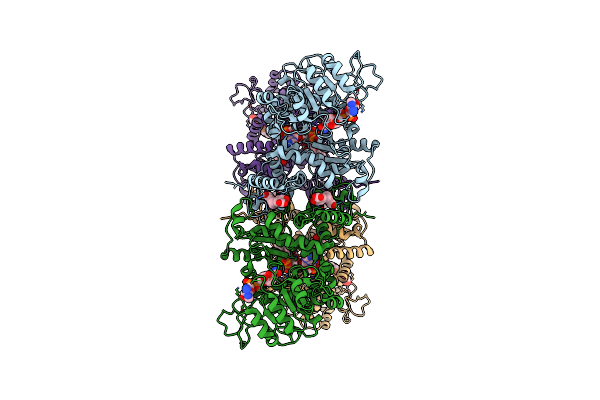 |
Cryo-Em Structures Of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme In A Ternary Complex With Nad+ And Allosteric Inhibitor Mdsa
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: HYDROLASE Ligands: NAD, D5S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: HMT |
 |
Organism: Edwardsiella piscicida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-20 Classification: CHAPERONE |
 |
Organism: Edwardsiella piscicida
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-07-20 Classification: CHAPERONE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-11-10 Classification: VIRAL PROTEIN Ligands: H3F |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: NAD, MG, FUM, TTN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: NAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: NAD |
 |
Crystal Structure Of Heptaprenyl Diphosphate Synthase From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2016-12-28 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Plasmodium Vivax Geranylgeranylpyrophosphate Synthase Complexed With Bph-1158
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-09-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 04M |
 |
Crystal Structure Of Plasmodium Vivax Geranylgeranylpyrophosphate Synthase Complexed With Bph-1182
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-09-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: HXK, SO4 |
 |
Crystal Structure Of Plasmodium Vivax Geranylgeranylpyrophosphate Synthase Complexed With Bph-1186
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2016-09-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 04W |
 |
Crystal Structure Of Plasmodium Vivax Geranylgeranylpyrophosphate Synthase Complexed With Bph-1251
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2016-09-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 63D |
 |
Crystal Structure Of Human Farnesyl Diphosphate Synthase In Complex With Bph-1358
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-04-15 Classification: TRANSFERASE Ligands: 3F2, PO4 |
 |
T. Brucei Farnesyl Diphosphate Synthase Complexed With Homorisedronate Bph-6
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2015-04-15 Classification: TRANSFERASE Ligands: HRX, MG |
 |
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-04-15 Classification: TRANSFERASE Ligands: MG, RIS |
 |
T. Brucei Farnesyl Diphosphate Synthase Complexed With Bisphosphonate Bph-14
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-04-15 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3YQ, MG |

