Search Count: 56
 |
Organism: Caldicellulosiruptor sp.
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Crystal Structure Of Beta-Glucosidase Cabgl Mutant E163Q In Complex With Glucose
Organism: Caldicellulosiruptor sp.
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, BGC, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: APOPTOSIS Ligands: MG, PD |
 |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: ANTIVIRAL PROTEIN/RNA |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: ANTIVIRAL PROTEIN/RNA |
 |
Crystal Structure Of Cellulosomal Double-Dockerin Module Of Clo1313_0689 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-03 Classification: PROTEIN BINDING Ligands: CA |
 |
Solution Nmr Structure Of Cellulosomal Double-Dockerin Module Of Clo1313_0689 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2024-04-03 Classification: PROTEIN BINDING Ligands: CA |
 |
Structure Of The Plasmodium Falciparum 20S Proteasome Complexed With Inhibitor Tdi-8304
Organism: Plasmodium falciparum nf54
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: YRE |
 |
Structure Of The Plasmodium Falciparum 20S Proteasome Beta-6 A117D Mutant Complexed With Inhibitor Wlw-Vs
Organism: Plasmodium falciparum dd2
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 7F1 |
 |
Structure Of Human Constitutive 20S Proteasome Complexed With The Inhibitor Tdi-8304
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: YRE |
 |
Crystal Structure Of Rna Helicase From Saint Louis Encephalitis Virus And Discovery Of Its Inhibitors
Organism: Saint louis encephalitis virus
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: EDO |
 |
Clostridium Thermocellum Rna Polymerase Transcription Open Complex With Sigi1 And Its Promoter
Organism: Acetivibrio thermocellus dsm1313, Acetivibrio thermocellus, Acetivibrio thermocellus dsm 1313
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: TRANSCRIPTION/DNA Ligands: ZN, MG |
 |
Clostridium Thermocellum Rna Polymerase Transcription Open Complex With Sigi6 And Its Promoter
Organism: Acetivibrio thermocellus dsm 1313, Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-08-23 Classification: HYDROLASE Ligands: GOL, BGC |
 |
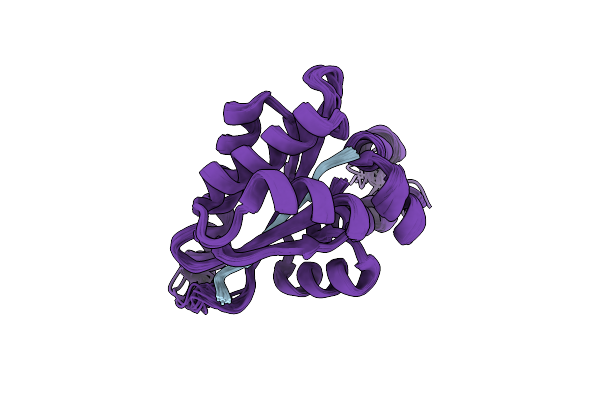
Solution Structure Of The Periplasmic Domain Of The Anti-Sigma Factor Rsgi1 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2023-05-24 Classification: SIGNALING PROTEIN |
 |
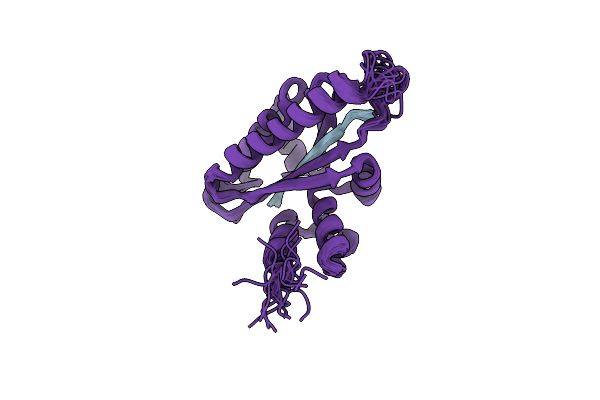
Solution Structure Of The Periplasmic Domain Of The Anti-Sigma Factor Rsgi2 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2023-05-24 Classification: SIGNALING PROTEIN |
 |
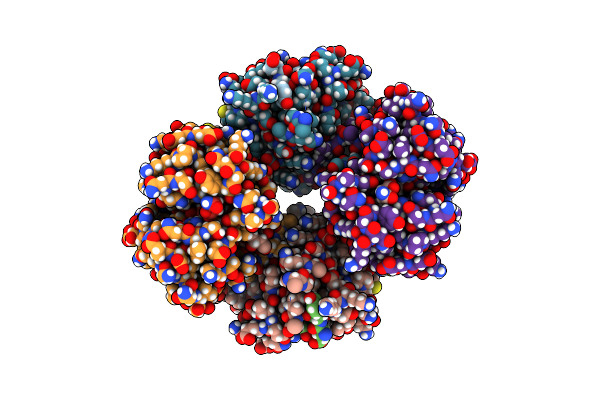
Solution Structure Of The Periplasmic Domain Of Rsgi6 From Clostridium Thermocellum
Organism: Acetivibrio thermocellus dsm 1313
Method: SOLUTION NMR Release Date: 2023-05-24 Classification: SIGNALING PROTEIN |
 |
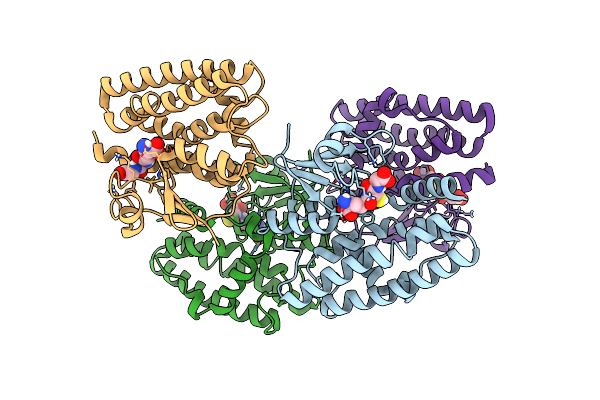
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-05-17 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of A Glutathione S-Transferase Tau1 From Pinus Densata In Complex With Gsh
Organism: Pinus densata
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-12-07 Classification: TRANSFERASE Ligands: GSH |

