Search Count: 18
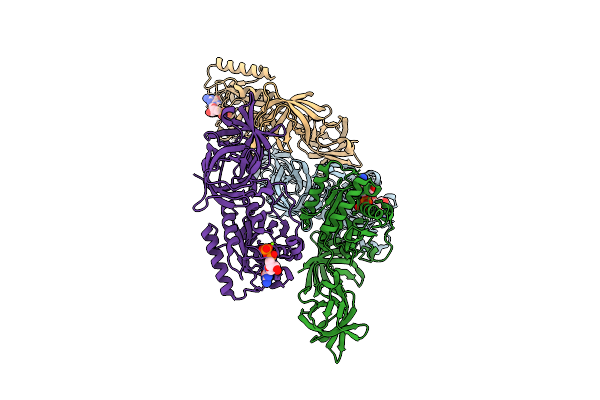 |
Organism: Mycoplasma pneumoniae (strain atcc 29342 / m129 / subtype 1)
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, MG |
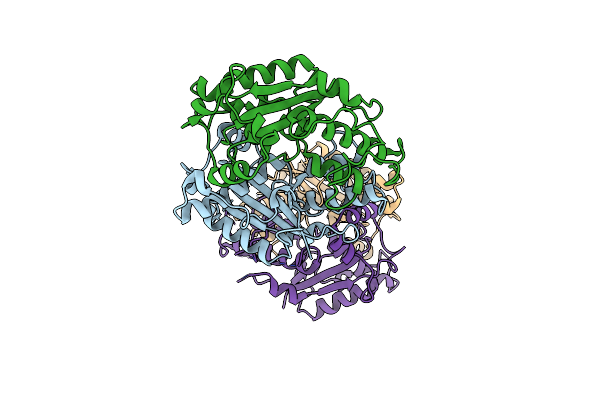 |
Organism: Escherichia coli 0.1197
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2025-02-19 Classification: DNA BINDING PROTEIN |
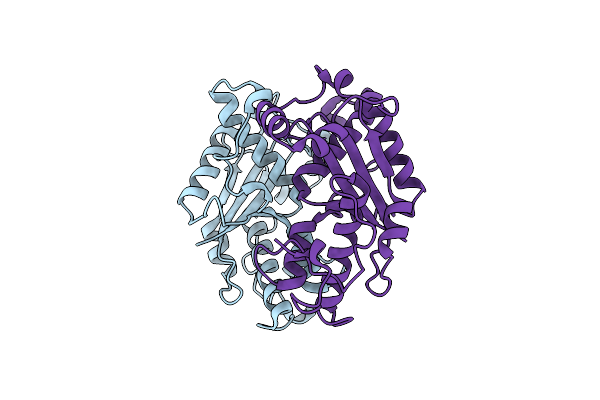 |
Organism: Escherichia phage ecszw_1
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2025-02-19 Classification: DNA BINDING PROTEIN |
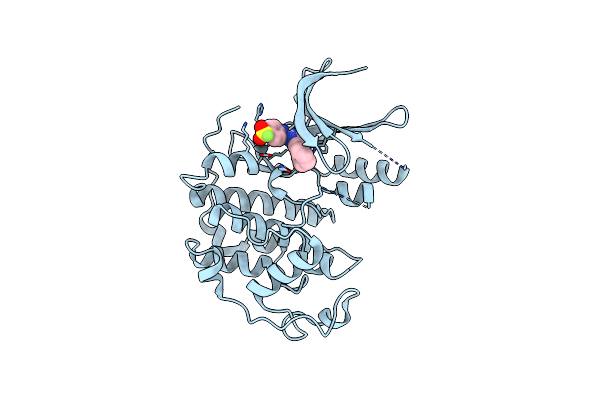 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-11 Classification: TRANSFERASE Ligands: A1IE9 |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2023-12-06 Classification: DNA BINDING PROTEIN |
 |
Organism: Pseudomonas phage jbd30, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-05-05 Classification: TRANSCRIPTION |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-04-12 Classification: HYDROLASE Ligands: PPI, BGC, GOL |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2017-04-12 Classification: HYDROLASE Ligands: BGC |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-04-12 Classification: HYDROLASE Ligands: BGC, GOL |
 |
Organism: Pseudomonas fluorescens (strain sbw25)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2017-01-25 Classification: UNKNOWN FUNCTION Ligands: EDO, CL, NA |
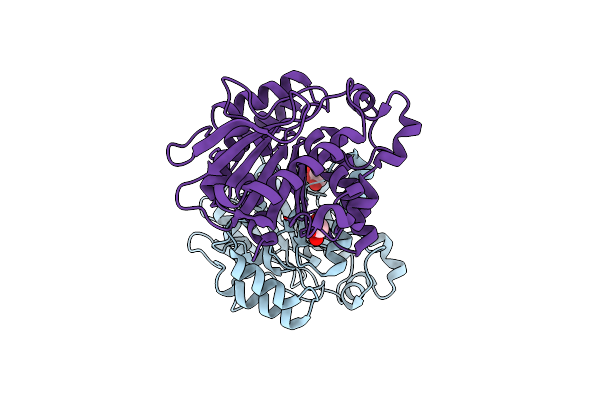 |
Crystal Structure Of A Cloned Feruloyl Esterase From A Soil Metagenomic Library
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-02-03 Classification: HYDROLASE Ligands: GOL |
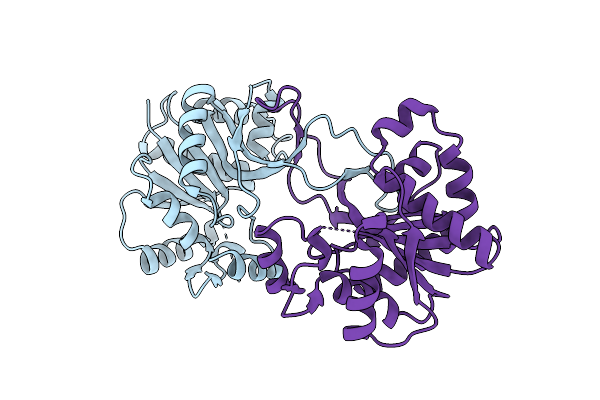 |
Crystal Structure Of Ribitol 5-Phosphate Cytidylyltransferase (Tari) From Bacillus Subtilis
Organism: Bacillus subtilis subsp. spizizenii
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-03-12 Classification: TRANSFERASE |
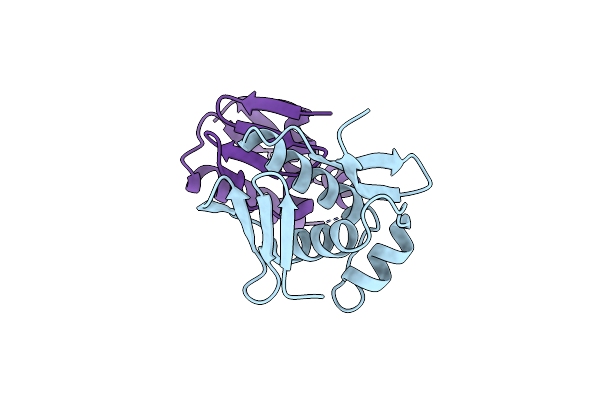 |
Structure Of Dna-Binding Domain Of The Response Regulator Saer From Staphylococcus Epidermidis
Organism: Staphylococcus epidermidis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-01-29 Classification: TRANSCRIPTION |
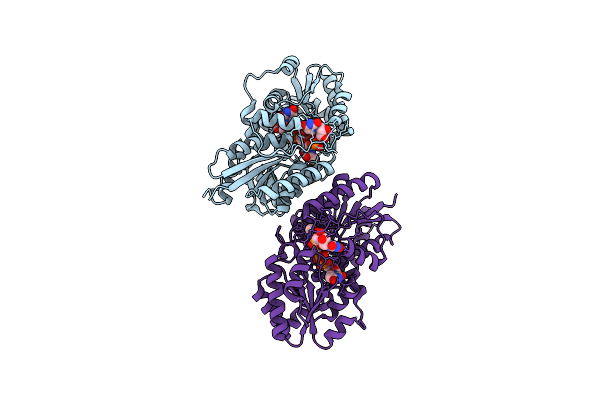 |
Crystal Structure Of Bacillus Subtilis Udp-Glcnac 2-Epimerase In Complex With Udp-Glcnac And Udp
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2013-05-08 Classification: BIOSYNTHETIC PROTEIN Ligands: UD1, UDP |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2012-07-11 Classification: TRANSFERASE Ligands: GOL, COA |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2012-07-11 Classification: ISOMERASE Ligands: PO4 |
 |
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-01-19 Classification: TRANSFERASE Ligands: NO3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-07-04 Classification: HYDROLASE Ligands: ZN, 541 |

