Search Count: 16
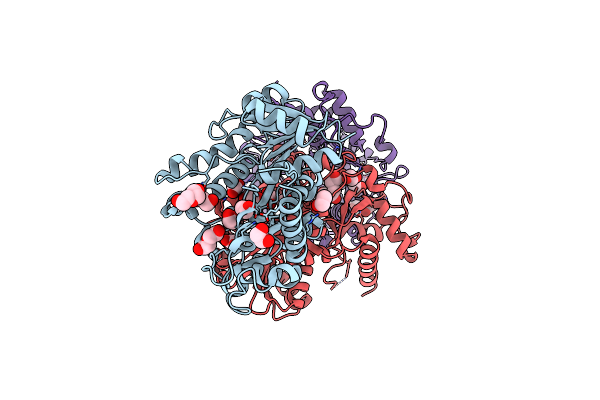 |
Organism: [candida] glabrata
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-08-02 Classification: OXIDOREDUCTASE Ligands: EDO, PEG, CIT |
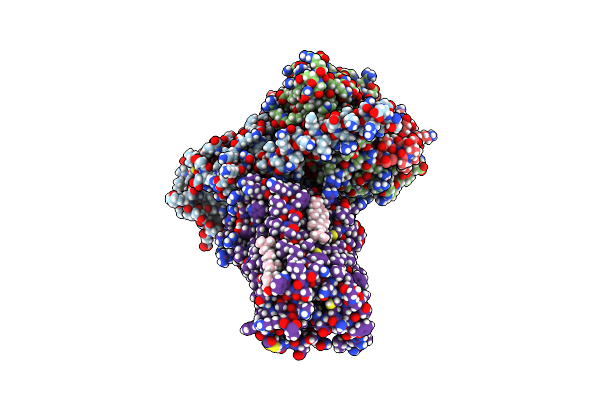 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: MEMBRANE PROTEIN Ligands: R5F, CLR |
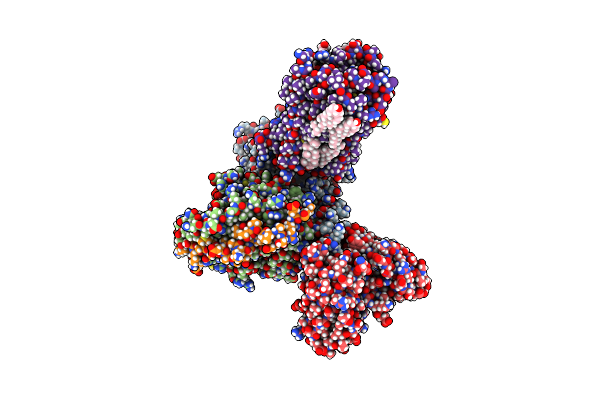 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: MEMBRANE PROTEIN Ligands: R5F, CLR |
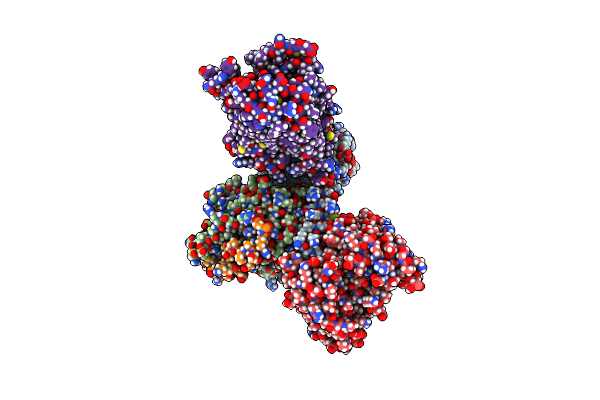 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: MEMBRANE PROTEIN Ligands: R5F |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: MEMBRANE PROTEIN Ligands: R5F |
 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: MEMBRANE PROTEIN Ligands: R5F, CLR, PLM |
 |
Cryo-Em Structure Of Skf-81297-Bound Dopamine Receptor 1 In Complex With Gs Protein
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: SK0, CLR, PLM |
 |
Cryo-Em Structure Of Skf-83959-Bound Dopamine Receptor 1 In Complex With Gs Protein
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: SK9, CLR, PLM |
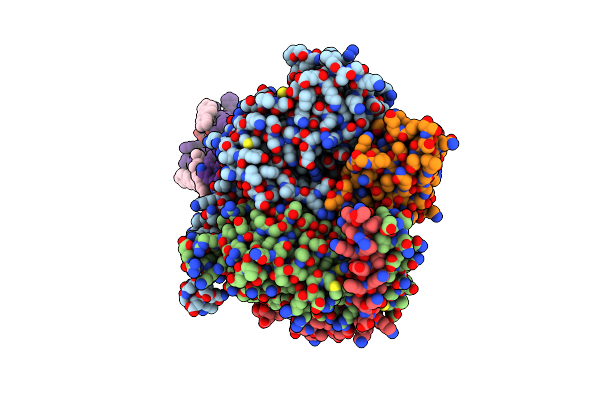 |
Cryo-Em Structure Of Apomorphine-Bound Dopamine Receptor 1 In Complex With Gs Protein
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: OR9, CLR, PLM |
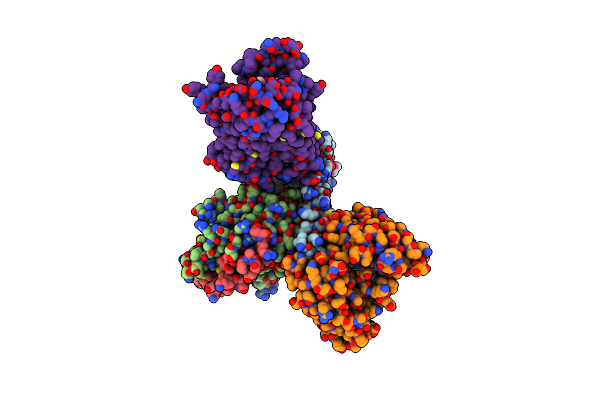 |
Cryo-Em Structure Of Bromocriptine-Bound Dopamine Receptor 2 In Complex With Gi Protein
Organism: Escherichia coli, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: SIGNALING PROTEIN Ligands: 08Y |
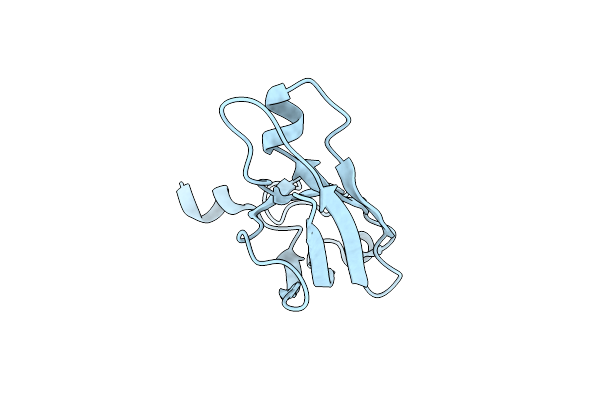 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2012-10-24 Classification: PROTEIN BINDING |
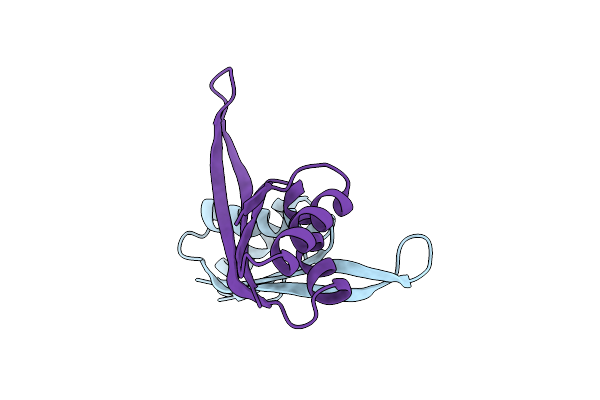 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-04-25 Classification: UNKNOWN FUNCTION |
 |
Organism: Lobophyllia hemprichii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-11-16 Classification: FLUORESCENT PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-05-25 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-03-09 Classification: DNA BINDING PROTEIN Ligands: MES, SO4 |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2007-05-08 Classification: HYDROLASE Ligands: NOL |

