Search Count: 1,731
 |
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Mus musculus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: 8WL |
 |
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Lama glama, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: DR7 |
 |
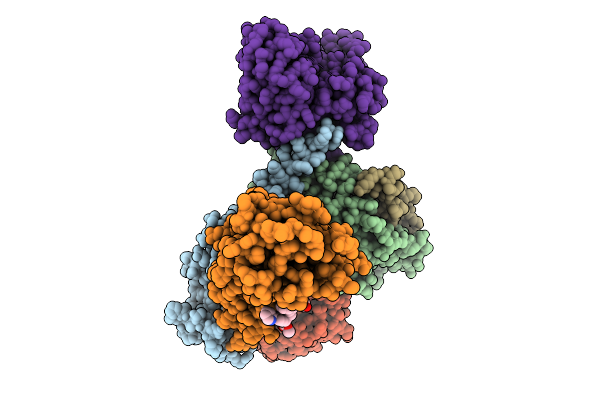
Structure Of Beta1-Ar-Gs Complex Bound To Epinephrine And An Allosteric Modulator
Organism: Bos taurus, Rattus norvegicus, Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: DR7, ALE |
 |
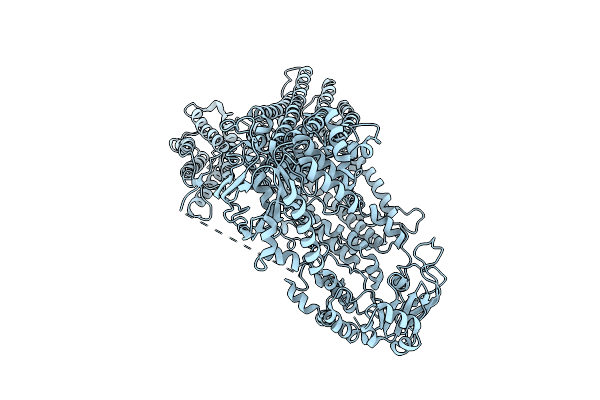
Organism: Bos taurus, Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: DR7, P0G |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN |
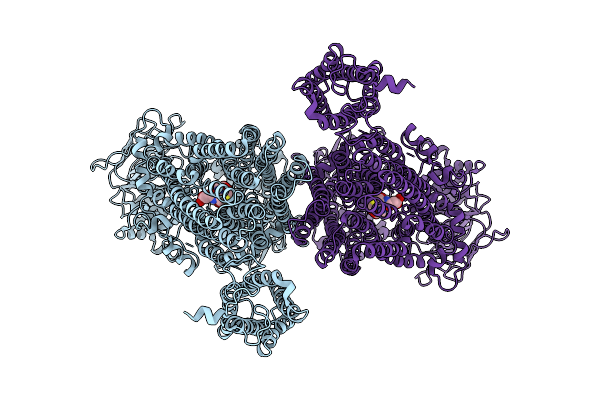 |
Structure Of Abcc2(E1404Q) In Arabidopsis Thaliana In The Dnp-Gs Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: GDN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
 |
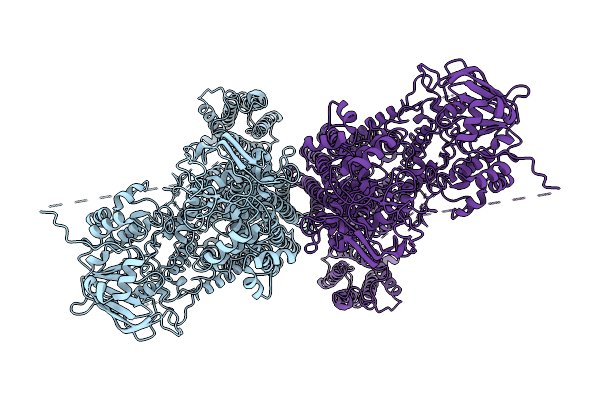
Structure Of Abcc2(E1404Q) Monomer In Arabidopsis Thaliana In The Dnp-Gs Bound State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: PROTEIN TRANSPORT Ligands: GDN |
 |
Structure Of The Wild-Type Abcc2 Dimer In Arabidopsis Thaliana In The Apo State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Organism: Burkholderia cenocepacia, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Xanthomonas, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: IMMUNE SYSTEM Ligands: GOL, PEG |
 |
Structure Of The S. Cerevisiae Clamp Loader Replication Factor C (Rfc) With Mixed Nucleotide Occupancy
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: REPLICATION Ligands: ACT, ADP, GDP |
 |
Structure Of The S. Cerevisiae Clamp Loader Replication Factor C (Rfc) With Mixed Nucleotide Occupancy
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: REPLICATION Ligands: ACT, ADP, GDP |
 |
Structure Of The S. Cerevisiae Clamp Loader Replication Factor C (Rfc) With Mixed Nucleotide Occupancy
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: REPLICATION Ligands: ACT, ADP, GDP |

