Search Count: 352
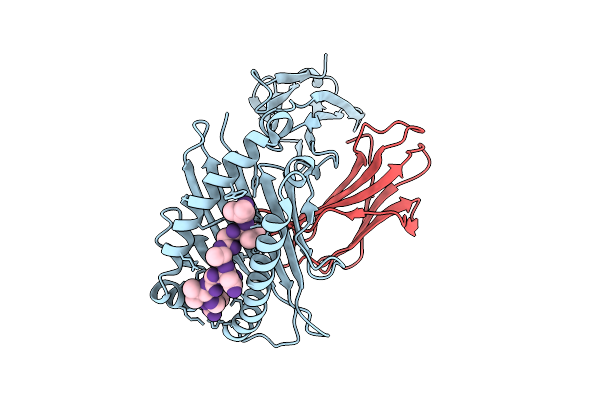 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12D 9-Mer Peptide (Vvgadgvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12A 10-Mer Peptide (Vvvgaagvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12S 10-Mer Peptide (Vvvgasgvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOKINE/IMMUNE SYSTEM |
 |
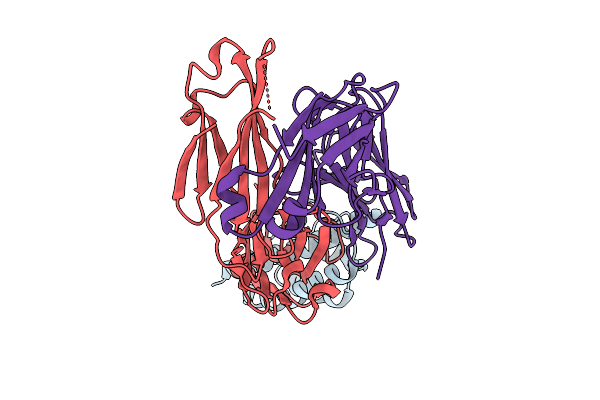
Structural Insights Into Il-31 Signaling Inhibition By A Neutralization Antibody
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOKINE/IMMUNE SYSTEM |
 |
Solution Structure Of Staphylococcus Aureus Response Regulator Arlr Dna-Binding Domain
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: SOLUTION NMR Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN |
 |
Structure Of The Rattus Norvegicus Ace2 Receptor Bound Hsitaly2011 Rbd Complex
Organism: Rattus norvegicus, Merbecovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG, ZN |
 |
Organism: Eptesicus fuscus, Merbecovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG, ZN |
 |
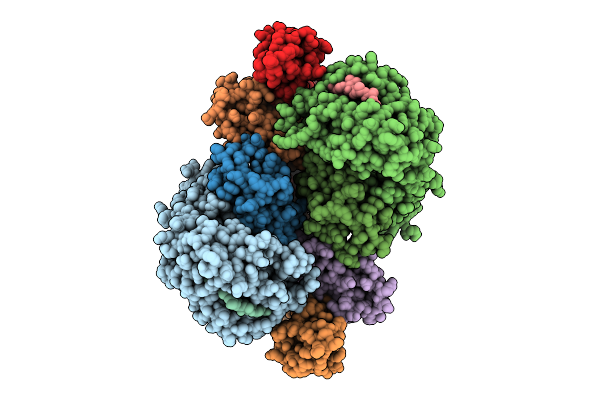
Fadd-Ded Filaments Coordinate Complex Iia Assembly During Tnf-Induced Apoptosis
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM |
 |
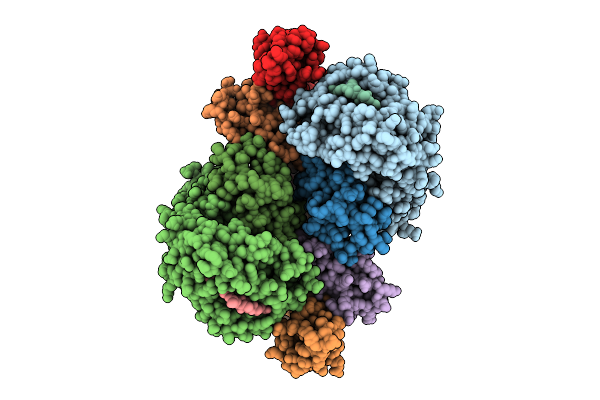
Fadd-Ded Filaments Coordinate Complex Iia Assembly During Tnf-Induced Apoptosis
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-05-07 Classification: DE NOVO PROTEIN Ligands: WUP, CL |
 |
Organism: Respiratory syncytial virus a2, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: HOH |
 |
Crystal Structure Of N-Acyl Homoserine Lactonase Ahlx Mutant M41(E77I/D177G/T243Y/H255L)
Organism: Salinicola salarius
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: NI |
 |
Organism: Salinicola salarius
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: NI, MG |
 |
The Binary Complex Structure Of F2Y224-Ftmox1 Mutant With Alpha-Ketoglutarate
Organism: Aspergillus fumigatus af293
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: CO, AKG, GOL, SO4 |
 |
Organism: Aspergillus fumigatus af293
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FE2, CO |
 |
Organism: Merbecovirus, Pteronotus davyi
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2025-02-26 Classification: VIRAL PROTEIN Ligands: NAG, ZN, FOL, EIC |
 |
Organism: Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, FOL, EIC |
 |
Organism: Pipistrellus abramus, Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-02-19 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG, ZN |

